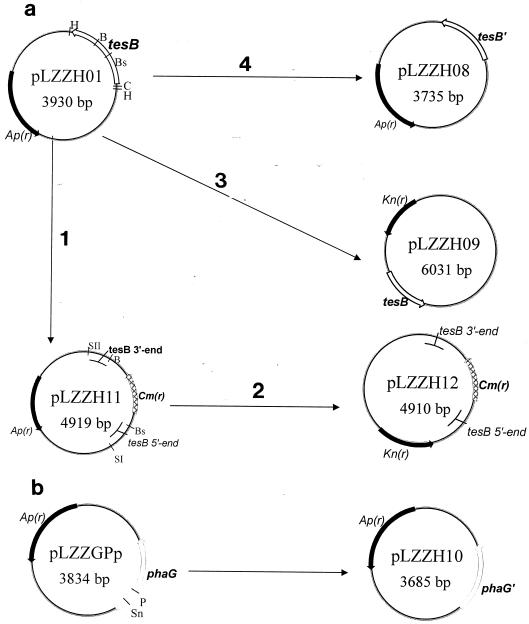
FIG. 1.
Scheme of plasmids used in this study. (a) The tesB gene from the E. coli JM105 genome was amplified by PCR with primers P1 and P3 and inserted into the pGEM-T vector to yield the starting plasmid, pLZZH01. The chloramphenicol resistance cassette from pBBR1MCS replaced the BstBI/BamHI fragment of tesB in pLZZH01, leading to the plasmid pLZZH11 (step 1). The SacII (filled in)-SacI fragment from pLZZH11 was inserted into the HincII/SacI sites of the temperature-sensitive vector pTH19ks1, resulting in pLZZH12 (step 2), which was used to construct the tesB knockout mutant E. coli strain. The HincII/ClaI fragment of pLZZH01, containing the tesB gene, was introduced into the corresponding sites of the vector pBBR1MCS-2 to yield pLZZH09 (step 3). BstBI/BamHI double-digested pLZZH01 was sequentially blunt ended by T4 polymerase and ligated to obtain the competitive plasmid pLZZH08 for the tesB transcriptional assay (step 4). (b) pLZZGPp was digested with PmlI and SnaBI, followed by ligation, resulting in pLZZH10, used for the transcriptional assay for phaG. tesB encodes thioesterase II; tesB′, the BstBI/BamHI fragment of tesB, was deleted; phaG encodes 3HD-ACP-CoA transacylase; phaG′, the SnaBI/PmlI fragment of phaG, was deleted. Ap(r), Kn(r), and Cm(r), ampicillin, kanamycin, and chloramphenicol resistance genes, respectively. B, BamHI; Bs, BstBI; C, ClaI; H, HincII; P, PmlI; SI, SacI; SII, SacII; Sn, SnaBI.