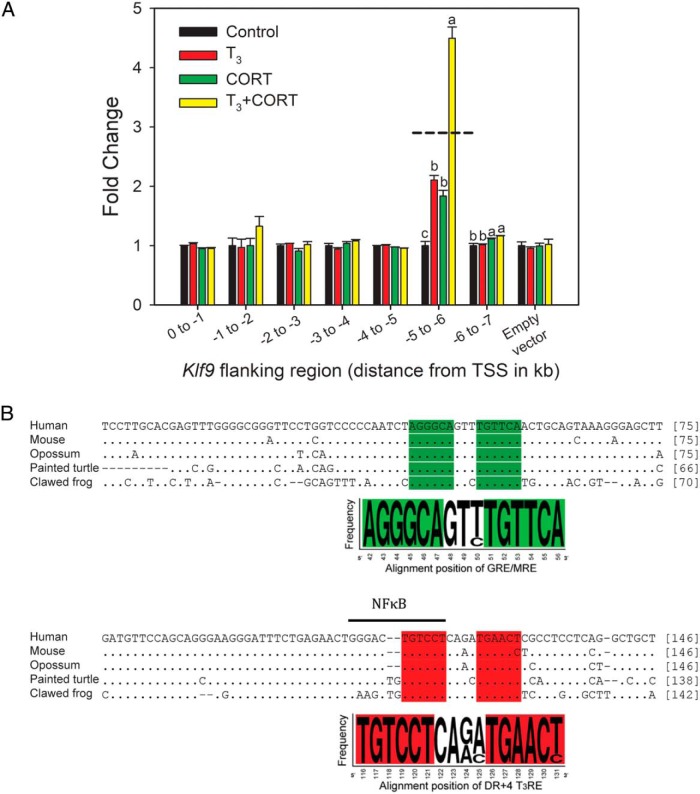
Figure 2.
Identification of an evolutionarily conserved approximately 180-bp genomic region upstream of tetrapod Klf9 genes that supports synergistic transactivation by TH and CORT. A, Enhancer-reporter scan of the 5′-flanking region of the X. tropicalis Klf9 gene; 1-kb fragments covering 0 to −7 kb upstream of the Klf9 TSS were cloned into the luciferase vector pGL4.23 and transfected into XTC-2 cells. We treated cells with vehicle (0.03% DMSO, 0.001% EtOH), T3 (5nM), CORT (100nM), or T3 plus CORT for 4 hours before harvest and analysis by dual luciferase assay. The −5- to −6-kb fragment supported individual hormone responses and hormone synergy (F(3,15) = 89.44, P < .001; ANOVA). The −6- to −7-kb fragment showed a small response to CORT (F(3,15) = 11.64, P < .05; ANOVA) that was unaffected by T3. Bars represent the mean ± SEM (n = 4–5/treatment) fold change, and letters above the means indicate significant differences among hormone treatments within a promoter fragment (means with the same letter are not significantly different; Tukey's multiple comparison test; P < .05). The dashed line indicates the calculated additive effect of combined hormone treatment. B, A genomic region spanning approximately 180 bp upstream of tetrapod Klf9 genes is evolutionarily conserved (the KSM). Shown is the most highly conserved region spanning 138–146 nt for 5 tetrapods (see Supplemental Tables 4 and 5 for more comprehensive analyses). The highlighted sequences show the highly conserved GRE/MRE and DR+4 T3RE regions. The evolutionarily conserved, predicted NFκB site that overlaps with the T3RE is indicated by a bar over the sequence. Note that there is a 2-base insertion in nonmammalian tetrapods. Genomic location of the KSM relative to the TSS for species for which it was possible to determine: X. tropicalis: −6000 to −5836 bp; mouse: −5333 to −5154 bp; human: −4634 to −4505.