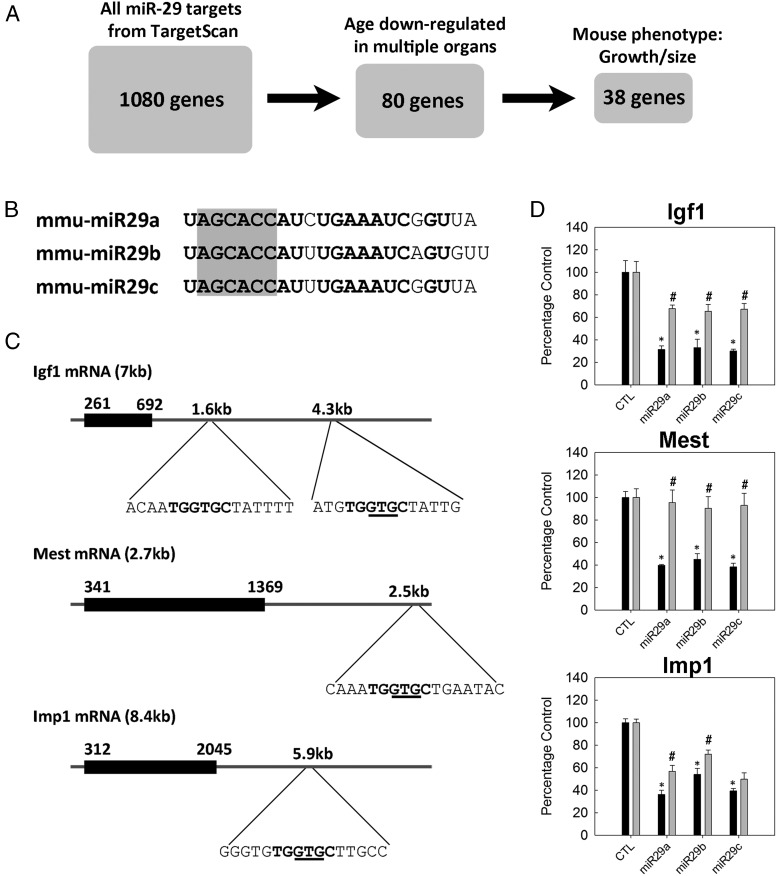
Figure 4.
In vitro validation of selected miR-29 target genes. A, A list of predicted miR-29 target genes that are relevant to our study was generated by filtering for down-regulation of expression with age in multiple organs and also for a phenotype involving body growth and/or body size in mice. B, Sequence of mouse miR-29a, -b, and -c. Seed regions are highlighted in gray. C, mRNA structures of mouse Igf1, Mest, and Imp1. Black boxes indicate coding regions; horizontal lines indicate UTRs. 3′-UTR regions containing predicted miR-29 target sites are expanded, with the sequence complementary to the seed regions shown in bold and the 3-nucleotides deleted by site-directed mutagenesis underlined. D, Igf1, Mest, or Imp1 3′-UTR regions were placed downstream from a luciferase reporter gene and then transfected, along with miR mimics, into HEK293 cells. miR-29a, -b, or -c mimics significantly reduced luciferase activity (black bars, means ± SEM) compared with that for a random mimic (CTL). *, P < .05 (ANOVA) compared with CTL. This reduction in luciferase activity was significantly diminished (gray bars) when 3 nucleotides were deleted from the miR-29 target region using site-directed mutagenesis. #, P < .05 (ANOVA) compared to the corresponding wild-type construct (adjacent black bar).