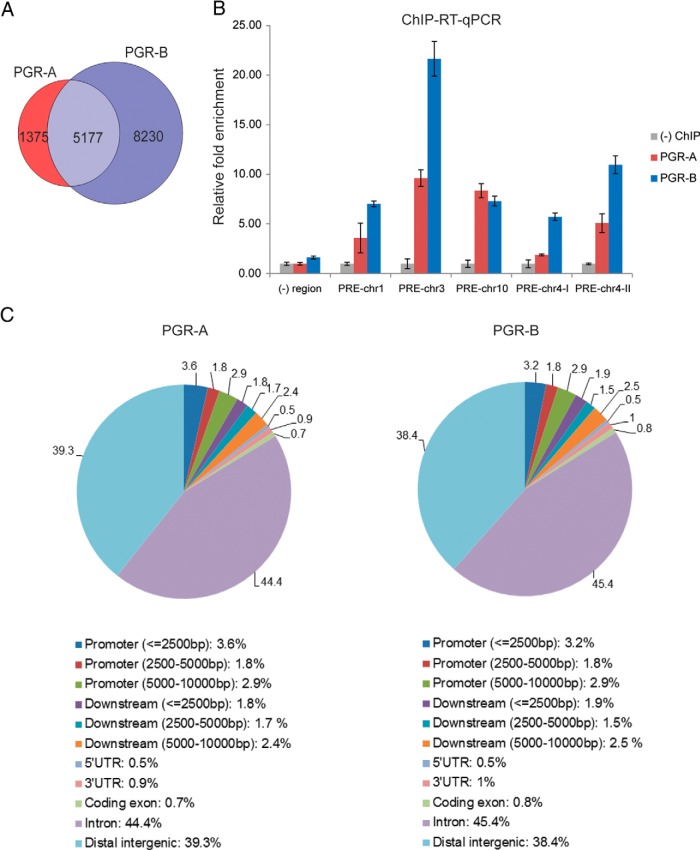
Figure 2.
Genome-wide PGR-A and PGR-B binding locations in differentiating HESCs. A, ChIP-seq–derived PGR-A and PGR-B binding sites. Venn diagrams show overlap of PGR-A vs PGR-B binding sites with a false discovery rate of <5% cutoff. B, A subset of PGR binding sites identified in the ChIP-seq experiment was selected for validation. ChIP was performed as described in Materials and Methods. Chromatin enrichment was quantified by real-time PCR using primers flanking these potential binding sites on different chromosomes as indicated in the figure. The negative (−) region was a PRE-deficient region. To calculate the enrichment of chromatin, the resulting signals were normalized to 1% input DNA. Data are presented as fold enrichment over that of the negative control ChIP and represent 3 independent experiments. C, Genomic region associations of genome-wide PGR-A and PGR-B binding sites. CEAS (a chromosome enrichment annotation tool of Cistrome) was used to analyze the distribution of binding sites with the following parameters: promoter/downstream lower interval, 2500 bp; middle interval, 5000 bp; and upper interval, 10 000 bp. In brief, regions between 0 and 2500 bp, 2500 and 5000 bp, and 5000 and 10 000 bp upstream of TSSs were selected as promoter intervals. Similarly, regions between 0 and 2500 bp, 2500 and 5000 bp, and 5000 and 10 000 bp downstream of a transcript were defined as different downstream intervals.