Figure 4.
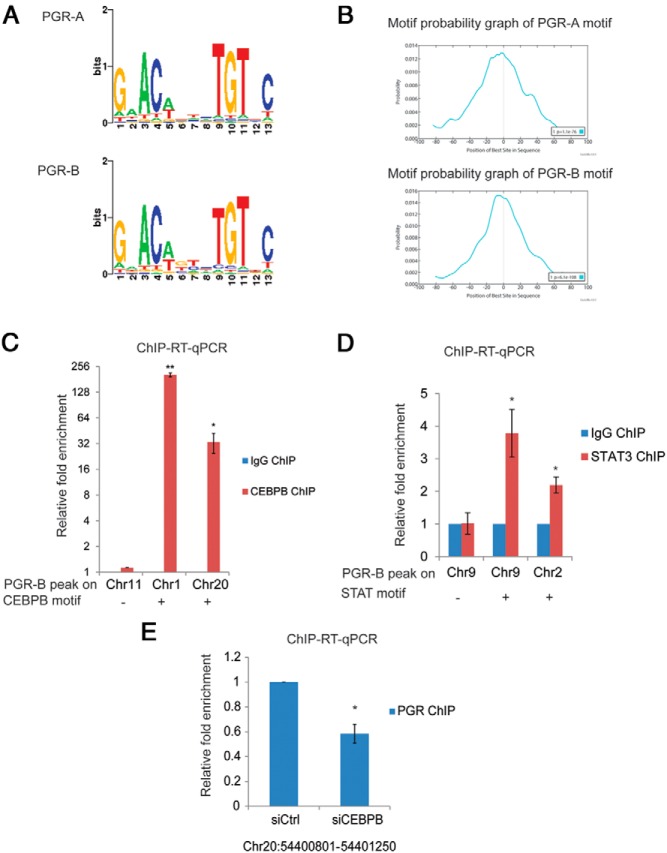
Motif analysis. A, PGR binding motif for PGR-A and PGR-B identified by de novo motif analysis using the SeqPos tool of Cistrome (22). B, Motif probability graphs showing the relative positions of PGR-A and PGR-B binding motifs within the summits of the top 1000 binding sites of PGR-A and PGR-B were generated using MEME-ChIP (21). The center of the sequences corresponds to the vertical line in the center of the graph. C, HESCs were subjected to treatment with differentiation cocktail for 48 hours. C/EBPβ ChIP was performed as described in Materials and Methods. Chromatin enrichment was quantified by real-time PCR using primers flanking the PGR-B binding sites, which contain a C/EBPβ motif within 100 bp (on chromosomes 1 and 20) and a C/EBPβ motif–negative PGR-B binding site (chromosome 11) as indicated in the figure. To calculate the enrichment of chromatin, the resulting signals were normalized to 1% input DNA. Data are presented as fold enrichment of C/EBPβ binding over that of the rabbit IgG ChIP. Graphs represent means ± SEM from 3 independent experiments. The Student t test was used for statistical analysis: **, P ≤ .01; *, P ≤ .05. D, HESCs were subjected to treatment with differentiation cocktail for 72 hours. STAT3 ChIP was performed as described in Materials and Methods. Chromatin enrichment was quantified by real-time PCR using primers flanking the PGR-B binding sites that contain a STAT motif within 100 bp (on chromosomes 9 and 2) and a STAT motif–negative PGR-B binding site (on chromosome 9) as indicated in the figure. To calculate the enrichment of chromatin, the resulting signals were normalized to 1% input DNA. Data are presented as fold enrichment of STAT3 binding over that of the IgG ChIP. Graphs represent the means ± SEM from 3 experiments. The Student t test was used for statistical analysis: *, P ≤ .05. E, HESCs were transfected with siRNA targeting C/EBPβ. At 48 hours after transfection, cells were treated with differentiation cocktail for 48 hours. PGR ChIP was performed. Chromatin enrichment was quantified by real-time PCR using primers flanking the PGR-B binding site on chr20: 54400801 to 54401250. To calculate the enrichment of chromatin, the resulting signals were normalized to 1% input DNA. Data are presented as fold enrichment of PGR enrichment in C/EBPβ siRNA-treated cells over that in control siRNA-treated cells. Graphs represent means ± SEM from 3 independent experiments. The Student t test was used for statistical analysis: *, P ≤ .05.