Figure 2.
Model of a Nonrandom Genetic Landscape in Gliomas: Discovery Set
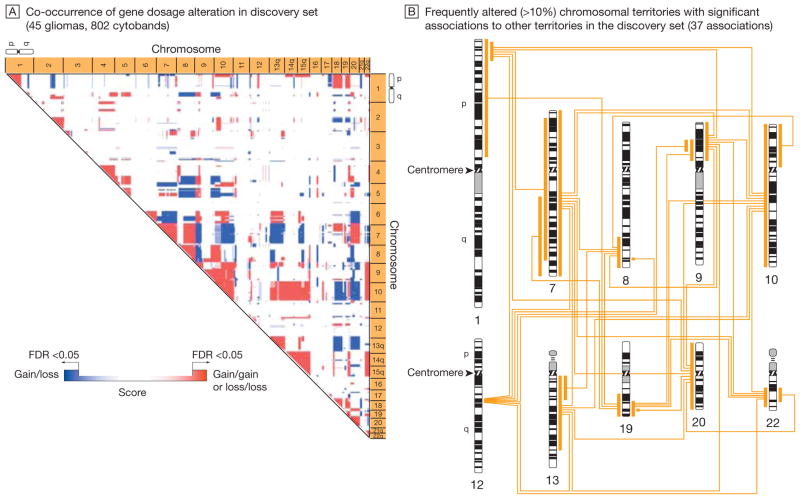
A permutation-based approach, which calculates the probabilistic fit for the coincidence of distinct chromosomal alterations, was applied to the gene dosage data in the discovery set from Stanford University. The gene dosage data generated by the circular binary segmentation change point algorithm were averaged according to 802 cytogenetic bands (cytobands)—subregions of a chromosome visible microscopically after special staining—that correspond to an International System for Human Cytogenetic Nomenclature (ISCN) 850 chromosome ideogram. The association map (A) indicates chromosomal regions (territories) with significant co-occurrence of gene dosage alteration (false-discovery rate [FDR]–corrected P<.05). Red scores denote the significant association of 2 chromosomal gains or 2 chromosomal losses; blue scores, the significant association of a gain and a loss event. The color gradation reflects a score that indicates the significance of cytoband-cytoband associations. The association map shows a consistent pattern of significant associations, which denotes a nonrandom genetic landscape in gliomas. B, Chromosomal territories that showed an alteration frequency of greater than 10% (involving chromosomes 1p, 7, 8q, 9p, 10, 12, 13, 19, 20, and 22) in the discovery set and their significant associations are mapped to a human ISCN-850 chromosome ideogram. (See interactive Figure 1 showing significant associations in the discovery and validation sets at http://www.jama.com.)