Figure 2.
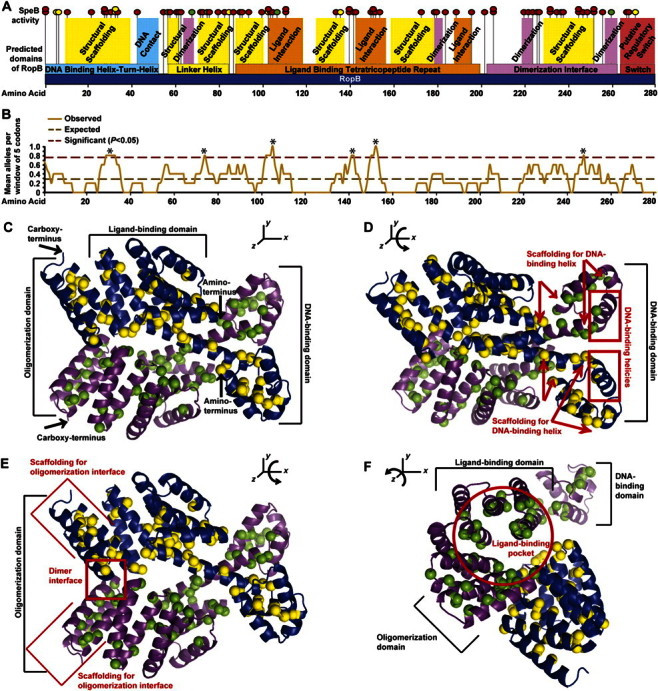
The ropB polymorphisms are predicted to alter RopB function. A, Location of each ropB polymorphism (vertical black lines with attached spheres) was mapped to the protein sequence (horizontal dark blue box with amino acid positions labeled). The DNA binding domain (horizontal light blue box), linker helix domain (horizontal yellow box), oligomerization domain (horizontal pink box), ligand interaction domain (horizontal orange box), and putative regulatory switch (horizontal red box) are shown. Protein regions predicted to form the DNA binding helix (vertical light blue box), ligand interaction interface (vertical orange box), oligomerization interface (vertical pink box) and structural scaffolding for these key functional domains (vertical yellow boxes) are also shown. The effect of each ropB polymoriphism on secreted streptococcal cysteine protease B (SpeB) expression and activity is also indicated (red, yellow, and green circles placed on top of each vertical line denoting the location of a particular polymorphism indicate absent, reduced or near wild-type levels of SpeB, respectively). The 11 codons altered by 2 different genetic changes in 2 different strains (eg, R28K and R28G) are indicated by partially overlapping stacked circles. B, Moving average plot of ropB polymorphisms per codon using a window of 5 amino acid residues (solid light brown line) was generated. Assuming a random distribution of altered codons, a rate of 0.29 polymorphisms per residue was expected (dashed dark brown line). Six regions with a significantly increased concentration of alterations were identified by the moving window analysis (asterisk above dashed maroon line indicates >0.77 polymorphisms per residue; χ2 test, P < .05). C, Structural model of the RopB dimer is shown, with the backbone of each monomer represented as a ribbon (violet and blue ribbons, respectively) and each amino acid altered by a polymorphism represented as a space filling sphere (green and yellow spheres, respectively). The DNA-binding domain, ligand-binding domain, oligomerization domain, and amino- and carboxy-terminus of each chain are labeled. D, Compared with view in C, the RopB structure has been rotated ∼45° around the x-axis to show the DNA-binding domain. Many amino acid replacements occur in the scaffolding helices (boundaries denoted by red arrows), but none occur in the DNA-binding helix (red box). E, Compared with view in C, the RopB structure has been rotated ∼25° around the x-axis to show the oligomerization domain. Many amino acid replacements occur in both the dimer interface (red box) and the scaffolding helices (boundaries denoted by red brackets). F, Compared with view in C, the RopB structure has been rotated ∼75° around the z-axis and ∼25° around the y-axis to show the ligand-binding domain. Note that many amino acid replacements occur along the interior surface of the ligand-binding pocket (red circle).
