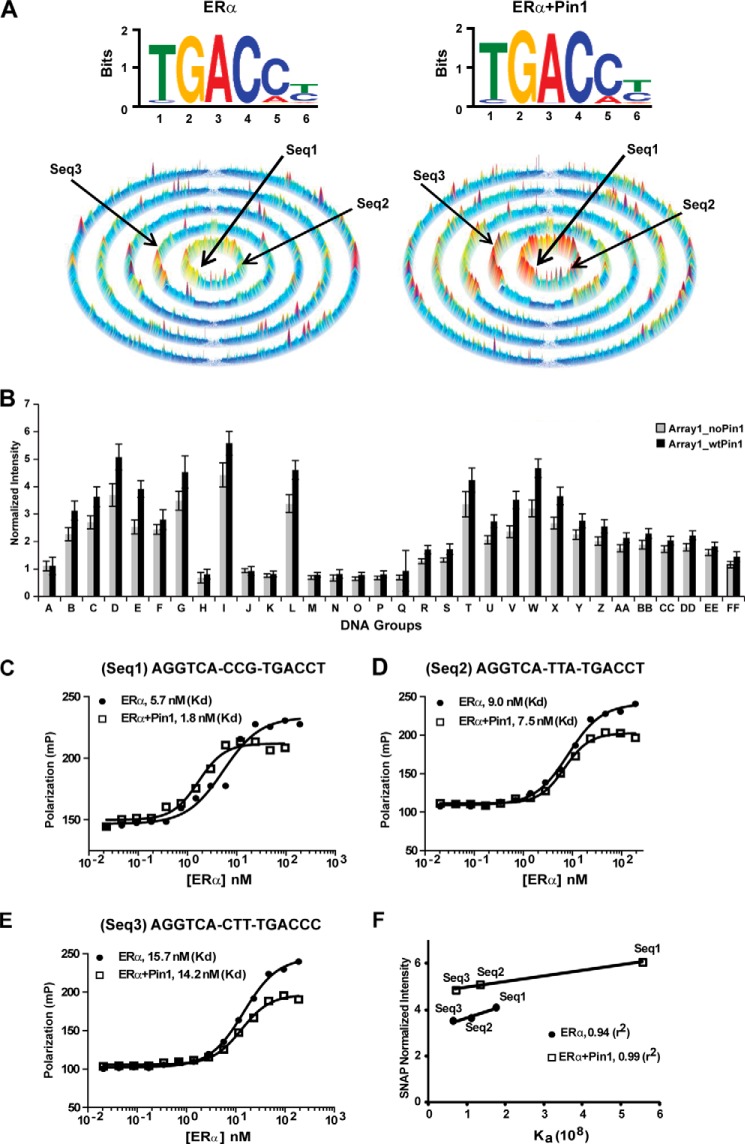
FIGURE 5.
Effect of Pin1 on ERα DNA binding using the comprehensive SNAP microarray. A, top, motif for estrogen-occupied ERα binding to all permutations of the core sequence, AGGTCAN9, in the 25-bp hairpin on the SNAP array. Bottom, SSL for estrogen-occupied ERα binding to all 262,144 (9-mer) DNA sequences. Each ring contains sequences with increasing mismatches from the seed motif composed of the sequences NNNTGACCT and TGACCTNNN. Left, motif and SSL for ERα. Right, motif and SSL for ERα + Pin1. B, the effect of Pin1 was determined by plotting normalized fluorescence intensity for ERα (Array1_noPin1) versus ERα + Pin1 (Array1_wtPin1) binding to different DNA probes from the groups listed in Table 2. A representative graph from two independent replicates is shown. Error bars, 1 S.D. C–E, determination of Kd values for estrogen-occupied ERα (filled circles) or ERα + Pin1 (open squares) for three DNA sequences (Seq1, Seq2, and Seq3; shown by arrows in Fig. 5A) from the SNAP array using FP. F, correlation between normalized SNAP intensity and Ka, relating fluorescence intensities on the SNAP arrays to affinity of ERα (filled circles) or ERα + Pin1 (open squares) for each DNA sequence.