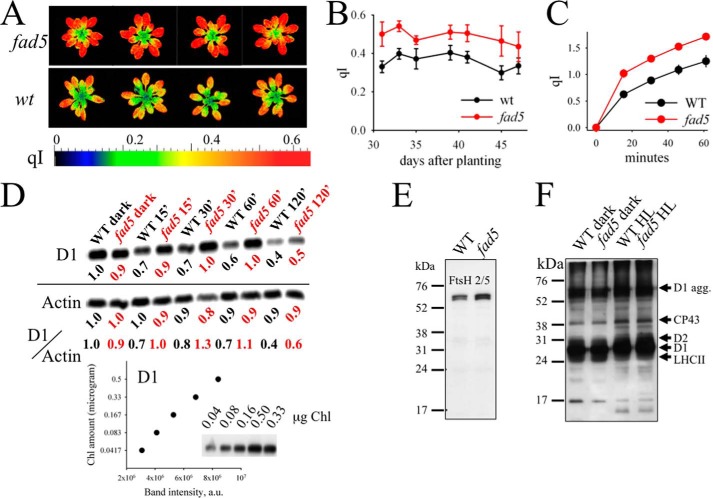
FIGURE 10.
fad5 mutant shows higher photoinhibition and an impaired PSII repair cycle. A, false color images for the qI parameter for fad5 and WT plants. B, qI parameter for whole plants determined as a function of time. Data represent the mean with standard error of 9 (fad5) and 18 (WT) plants. C, photoinhibition of isolated protoplasts induced by white light of 2000 μmol quanta m−2 s−1 at room temperature. For qI determination at different time points, strong light was switched off for 3 min and Fv′ was determined after this time. This procedure allows elimination of the reversible qE component that relaxes in a few minutes. Data represent the mean with standard error of 10 (WT) and 11 (fad5) samples. D, D1 dilution series and HL-induced changes in D1 content at 15, 30, 60, and 120 min of HL. The Western blots are representative of three independent analyses. The time indicates the duration of the strong light (1200 μmol quanta m−2 s−1) that induces PSII photoinhibition. All the time series samples have been treated with 1 mg/ml lincomycin and were loaded on the gel on an equal Chl basis (0.167 μg of Chl/sample). The loading control actin was detected in a similar gel with a higher sample concentration (1.0 μg of Chl/sample). The band intensities of D1 and actin, normalized to respective WT dark samples, are given below each lane. To account for the separate D1 and actin blots, D1 to actin ratios were also shown. The graph at the bottom shows the nearly linear correlation between D1 amount and the band intensity. E, Western blot analysis of the FtsH protein in the dark-adapted leaves. FtsH level is slightly elevated in the fad5 mutant. F, PSII-core phosphorylation level of dark and HL-treated samples. The antibody is directed against the phosphothreonine. The HL-induced increase in phosphorylation levels of D1, D2, and CP43 is well documented and results from the activation of the STN8 kinase. No differences in phosphorylation are visible between WT and fad5 mutant.