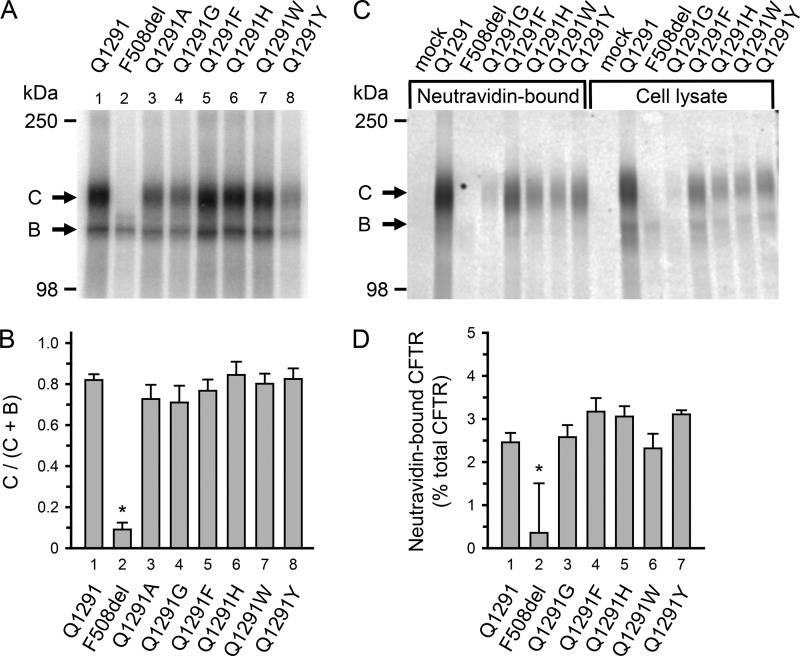
FIGURE 2.
Processing of CFTR with Gln-1291 mutations. A and B, processing was evaluated by assessing CFTR glycosylation. CFTR core glycosylation occurs in the endoplasmic reticulum. The core-glycosylated CFTR migrates as band B during SDS-gel electrophoresis. Later, CFTR becomes highly glycosylated in the Golgi apparatus and then migrates as band C. Deletion of phenylalanine 508 (F508del) illustrates a mutation causing a processing defect at 37 °C; most of F508del CFTR is degraded before reaching the Golgi complex and therefore does not appear as band C (31, 35, 87, 88). A, autoradiograph. Wild-type and mutant CFTR were expressed in 293T cells for 48 h at 37 °C using a vaccinia virus-T7 hybrid expression system (36). CFTR was solubilized, immunoprecipitated, and phosphorylated with the catalytic subunit of protein kinase A and [γ-32P]ATP (38). Immunoprecipitates were fractionated on 6% SDS-polyacrylamide gels. B, quantitative data for the fraction of CFTR migrating as band C. Radioactivity incorporated into bands B and C was quantified by digital autoradiography. Depicted is the ratio of radioactivity in band C versus the total radioactivity in bands B and C. Data for wild-type (Q1291) CFTR and F508del CFTR are depicted for comparison and have been previously shown in the supplemental material of Ref. 17. *, p < 0.001 when compared with bar 1, 3, 4, 5, 6, 7, or 8. No significant differences were detected between bars 1, 3, 4, 5, 6, 7, and 8 (one-way ANOVA followed by Holm-Sidak's method of all pairwise multiple comparisons; wild-type CFTR, n = 13; F508del CFTR, n = 10; Q1291A CFTR, n = 4; Q1291G CFTR, n = 4; Q1291F CFTR, n = 6; Q1291H CFTR, n = 6; Q1291W CFTR, n = 6; Q1291Y CFTR, n = 6). C and D, processing to the plasma membrane was evaluated by assessing CFTR cell surface biotinylation. Experiments were performed as described under “Experimental Procedures.” C, Western blot probed with CFTR antibody 769 of NeutrAvidin-bound protein from 97.5% of the total soluble cell lysate (left eight lanes) and 2.5% of the total cell lysate (before incubating with NeutrAvidin; right eight lanes). Letters label highly glycosylated (C) and core-glycosylated (B) CFTR. The NeutrAvidin-bound CFTR migrated almost exclusively as band C. Mock-transfected cells received pcDNATM3.1 vector without CFTR cDNA insert. D, quantitative data for the fraction of CFTR (in percent) that bound to NeutrAvidin after exposing intact cells to cell surface biotinylation. CFTR protein was quantified by Western blotting as described under “Experimental Procedures.” *, p < 0.01 when compared with bar 1, 3, 4, 5, 6, or 7 (one-way ANOVA followed by Holm-Sidak's method of multiple comparisons versus control group; wild-type CFTR, n = 6; F508del CFTR, n = 4; Q1291G CFTR, n = 4; Q1291F CFTR, n = 6; Q1291H CFTR, n = 4; Q1291W CFTR, n = 4; Q1291Y CFTR, n = 4). No significant differences were detected between bars 1, 3, 4, 5, 6, and 7 (one-way ANOVA). Error bars, S.E.