Figure 3.
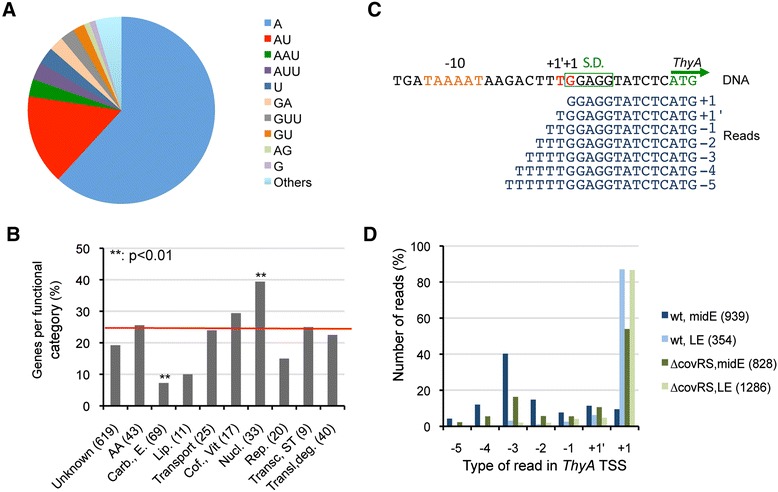
Extensive reiterative transcription in S. agalactiae. A. Pie chart of the occurrence of pseudo-templated nucleotides at the transcription initiation sites. B. Functional classification of genes submitted to reiterative transcription according to the KEGG categories (AA: Amino acid metabolism; Carb.,E.: carbohydrate, glycan and energy metabolism; Lip.: Lipid metabolism; Transport: Membrane transport; Cof.,Vit: metabolism of cofactors and vitamins; Nucl.: Nucleotide metabolism; Rep.: Replication and Repair; Transc, ST: Transcription, Signal transduction; Transl., deg.: translation, protein folding, sorting and degradation; Unknown: genes for which the KEGG category was not defined). TSSs were classified according to the functional class of the first gene of the transcription unit. The number of TSSs linked to each class is indicated in brackets. The mean proportion of TSSs with reiterative transcription is indicated by the red line. The proportion of genes showing reiterative transcription was found to be significantly higher (p < 0.01) for genes involved in nucleotide metabolism and lower (p < 0.01) for genes involved in carbohydrate, glycan and energy metabolism using chi-square and Fisher exact tests. C. Reiterative transcription at the thyA gene. The promoter of thyA directs initiation of transcription at a T or a G (shown in red on the non-template strand of the DNA sequence). The G position also corresponds to the first nucleotide of the Shine-Dalgarno (SD) sequence. Initiation at the T position leads to reiterative incorporation of pseudo-templated U. D. The proportion of reads containing pseudo-templated nucleotides in thyA transcripts is found to vary depending on growth conditions: mid-exponential and late exponential phases in the wt and the ∆covRS mutant of strain NEM316. The total number of reads corresponding to the TSS is indicated in brackets next to the experimental condition in the figure legend.
