Figure 5.
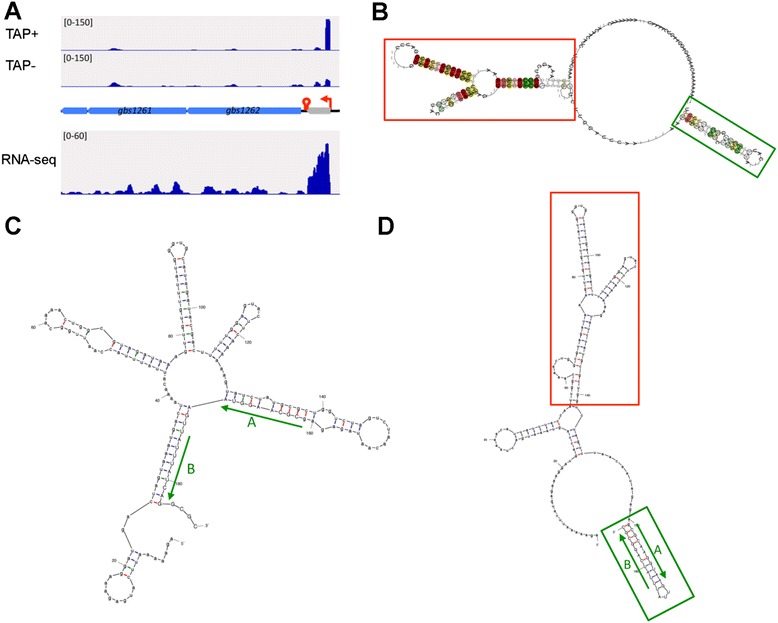
Identification of a novel riboswitch upstream gbs1262. A . Transcriptional organization deduced from dRNA-seq and RNA-seq experiments. Reads aligning upstream gbs1262 were visualized by IGV browser [67]. The sequence of the sRNA resulting from transcription premature arrest at a rho-independent terminator is indicated as a grey arrow. B. Secondary structure predicted by RNalifold [71], based on the alignment of 14 sequences similar to gbs1262 5′UTR in Lactobacillales and upstream a potential tryptophan-related gene in F. nucleatum (Additional file 9). The two conserved folded structures are indicated in red and green boxes, with the green box corresponding to a rho-independent terminator. Sequence covariations supporting the consensus structure are marked by color: red marks pairs with no sequence variation; ochre and green mark pairs with 2 or 3 types of pairs, respectively. C and D. Alternative structures of gbs1262 putative riboswitch as determined by mfold [72], showing the formation of an antiterminator structure.
