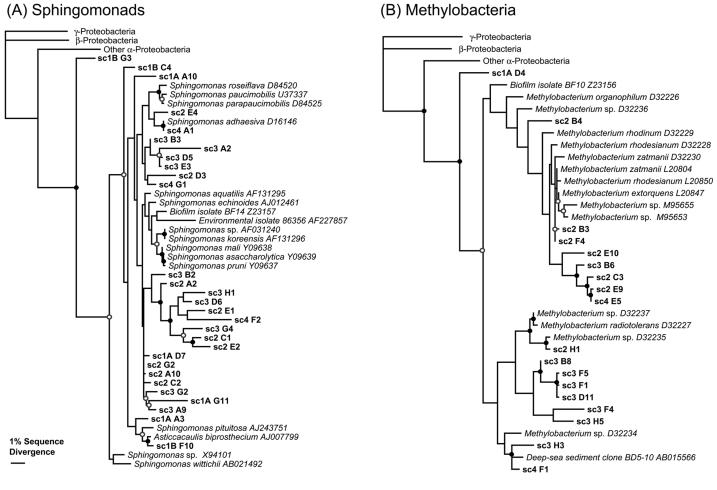
FIG. 2.
Results of phylogenetic analyses with Sphingomonas spp.-related (A) and Methylobacterium spp.-related (B) 16S rRNA sequences obtained in this study. The analyses performed were based on alignments of approximately 670 nucleotide positions. The alignments included cultured Sphingomonas and Methylobacterium spp. and outgroup γ-proteobacteria (Escherichia coli L10328), β-proteobacteria (Nitrosospira mutiformis L35509), and α-proteobacteria (Roseobacter denitrificans M59063). The phylogenetic trees shown were estimated by using ML. MP and NJ analyses converged on very similar tree topologies. Filled circles indicate both MP and NJ bootstrap support exceeding 70%, and open circles indicate bootstrap support exceeding 50%. (See Materials and Methods for details on the phylogenetic and bootstrap analyses.).