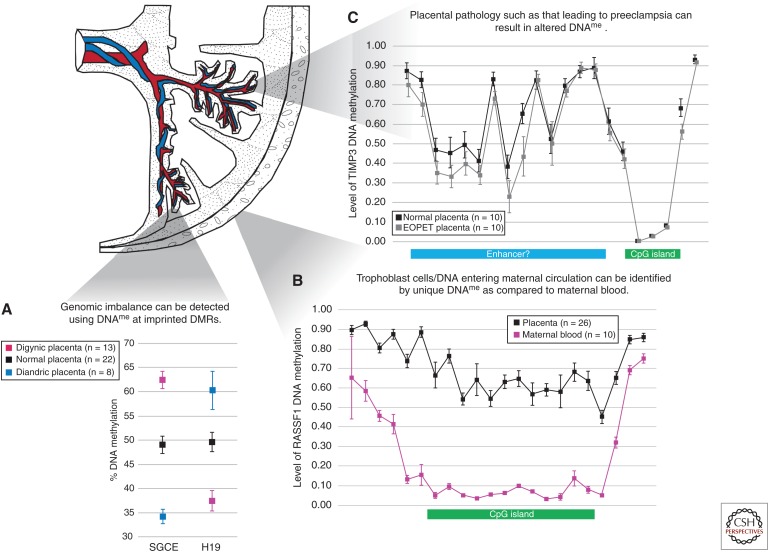
Figure 2.
Applications of placental DNA methylation. (A) DNA methylation at imprinting control regions (ICRs) such as those associated with the genes SGCE and H19 can be used to identify genomic imbalance in the placenta. Mean and standard deviation of DNA methylation levels for digynic and diandric triploidy and controls are indicated. (Adapted from Bourque et al. 2010.) (B) Regions of the genome with distinct patterns of placental DNA methylation as compared with that in maternal blood, for example at the CpG island of RASSF1 depicted here, can be used to distinguish fetal DNA in maternal circulation. Each data point represents the mean level of DNA methylation at a single CpG site obtained from the Illumina HumanMethylation450 BeadChip array; error bars represent standard deviation of the mean (WP Robinson and EM Price, unpubl.). (C) Altered DNA methylation may be detected in the placentas of complicated pregnancies, such as in early onset preeclampsia (EOPET). These may not be restricted to CpG islands; in TIMP3 (chr22: 33195342–33257640), for example, lower DNA methylation is observed in the putative enhancer region. Each data point represents the mean level of DNA methylation at a single CpG site obtained from the Illumina HumanMethylation450 BeadChip array; error bars represent standard deviation of the mean. (Adapted from Blair et al. 2013.) DNA methylation, DNAme.