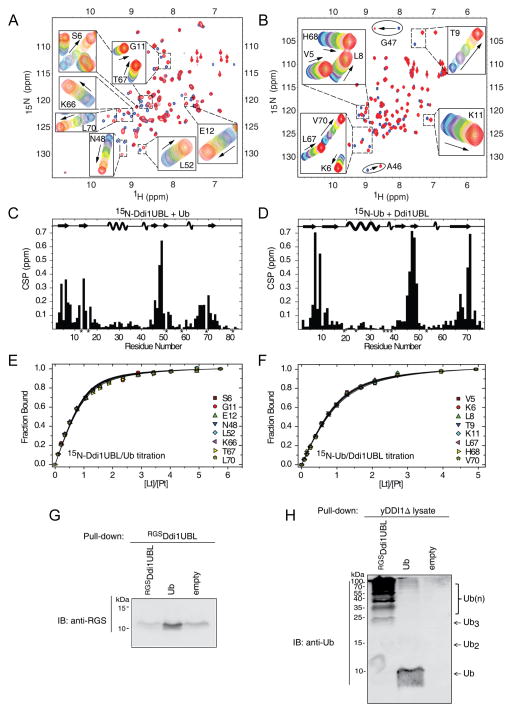
Figure 6.
NMR characterization of Ub binding to Ddi1UBL. (A) Overlay of 1H-15N SOFAST-HMQC spectra of 15N-labeled Ddi1UBL free in solution (blue) and in Ub-bound state (at Ub:Ddi1UBL molar ratio 5:1). Insets show zoom on selected regions to illustrate gradual shifts in the peak positions upon titration. (B) Overlay of 1H-15N SOFAST-HMQC spectra of 15N-labeled Ub alone (blue) and saturated (red) with Ddi1UBL (at 5-fold molar excess). Gradual signal shifts of the residues used for Kd estimation are shown in insets. A46 and G47 exhibiting slow exchange upon Ub binding are marked with ovals. (C) Amide CSPs in Ddi1UBL at the endpoint of titration with Ub (molar ratio 5.7:1) as a function of residue number. Asterisks indicate P16 and those residues where N-H resonances cannot be followed due to signal overlap. (D) CSPs in Ub at the endpoint of titration with Ddi1UBL (5:1 molar ratio). P19, P37, P38, as well as several amides that could not be reliably observed in the NMR spectra are marked with asterisks. The location of the secondary structure elements in Ddi1UBL and Ub is shown in the top portions of the graphs in (C) and (D), respectively. (E–F) Representative normalized titration curves for selected β-sheet residues of Ddi1UBL (E) or Ub (F). The curves show fits of the data to a 1:1 binding model. (G) Ub pulls down Ddi1UBL. Purified RGS-His6-Ddi1UBL or His6-Ub was immobilized to activated CH-sepharose beads and was incubated with RGS-His6-Ddi1UBL. Eluted analyte proteins were visualized by immunoblotting against RGS tag (see Supplemental Experimental Procedures for further details). (H) Ddi1UBL pulls out Ub conjugates from yeast cell extract. As in (G), purified RGS-His6-DDI1UBL or His6-Ub was attached to activated CH-sepharose beads. Total protein extract was obtained from growing DDI1Δ yeast cells (strain lacking Ddi1 gene). Beads loaded with proteins were incubated with cell extract. Eluted proteins were detected by immunoblotting with anti-Ub. The monomeric Ub seen in the elution from Ub-loaded beads likely reflects a small amount of the protein that came off the beads during elution with 8M urea. (See also Fig S7)