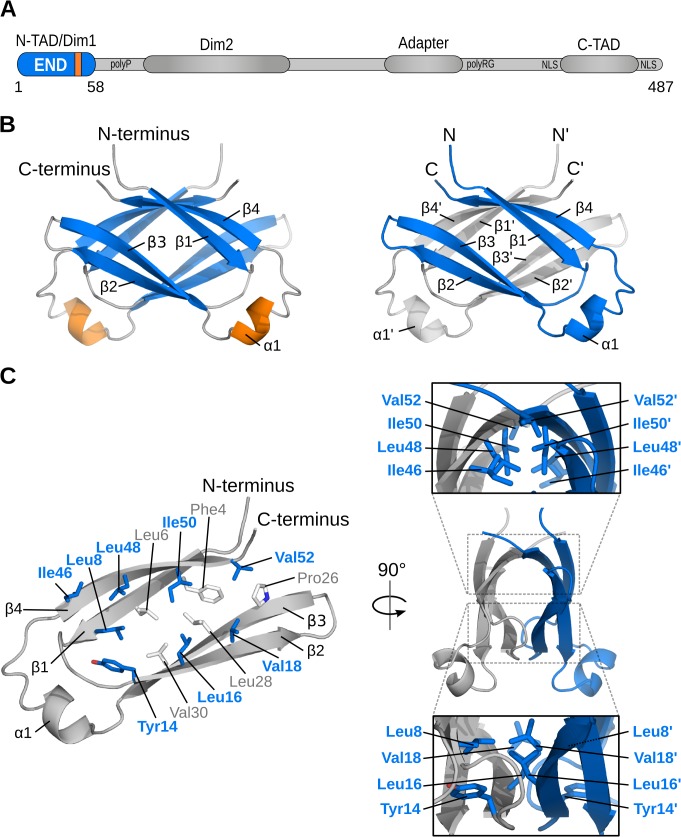
Fig 1. Structure of the EBNA-2 N-terminal dimerization (END) domain.
Schematic representation of important features of the EBNA-2 protein: two dimerization motifs (Dim1/Dim2), N-terminal and C-terminal transactivation domains (N-TAD, C-TAD), repetitive primary sequence motifs like the poly-proline (polyP) and the poly arginine-glycine (polyRG) stretch, the nuclear localization signals (NLS),and the adapter region of EBNA-2, which interacts with CBF1/CSL, are illustrated. (B) NMR solution structure of the END (EBNA-2 N-terminal Dimerization) domain. Left: β-strands are shown in blue, helices in orange, and loops in gray. Right: Monomers highlighted in gray and blue. (C) Dimerization of monomers is stabilized by hydrophobic interactions. The inside of each monomer is lined with numerous hydrophobic residues (left; sticks). A subset of these residues is located at the dimer interface (blue/bold labels). Panels (right) show side views of the END domain and highlight the interface residues of each monomer.