Figure 1. The M. tuberculosis DnaE1 polymerase encodes an intrinsic proofreading capability.
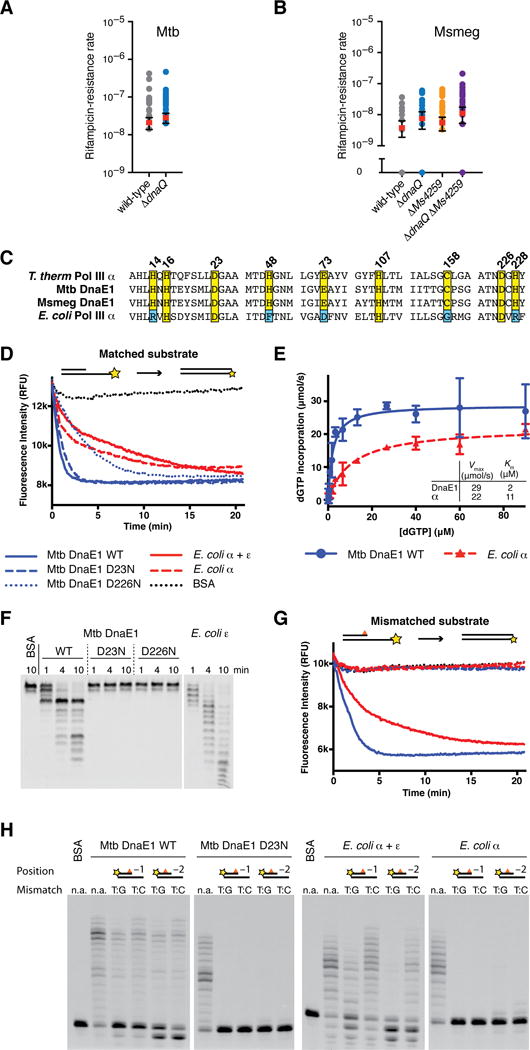
(A) Rates at which the indicated M. tuberculosis strains acquired resistance to rifampicin were measured by fluctuation analysis. Rv3711c is the annotated dnaQ gene. Circles represent mutant frequency (number of rifampicin-resistant mutants per cell plated in a single culture). Red bars represent the estimated mutation rates (mutations conferring rifampicin resistance per generation), with error bars representing the 95% confidence intervals.
(B) Fluctuation analysis was performed with the indicated M. smegmatis strains as in Figure 1A. Ms6275 is the annotated dnaQ gene and Ms4259 is next closest dnaQ homologue.
(C) Alignment of DNA polymerase PHP domains from the indicated species.
(D) Real-time primer extension activity of purified polymerases. Primer extension results in quenching of template fluorophore.
(E) Vmax and Km measurements derived from three primer extension assays. DnaE1MTB incorporates nucleotides faster than PolIIIαEC. Data points indicate the mean and error bars the standard deviation.
(F) Time course of 3′-5′ exonuclease activity on ssDNA. Wild-type DnaE1MTB shows robust exonuclease activity while the PHP mutants D23N & D226N do not. Note the distinct digestion patterns of DnaE1MTB and εEC-exonuclease.
(G) Primer extension assay as in Figure 2B with a mismatched DNA substrate. Exonuclease deficient polymerases cannot extend from mismatched DNA, while wild-type DnaE1MTB and PolIIIαEC+εEC activities are unaffected.
(H) Gel analysis of primer extension reactions shows that extension from mismatched primers requires exonuclease activity, while extension activity on matched substrates (n.a.) is unaffected.
