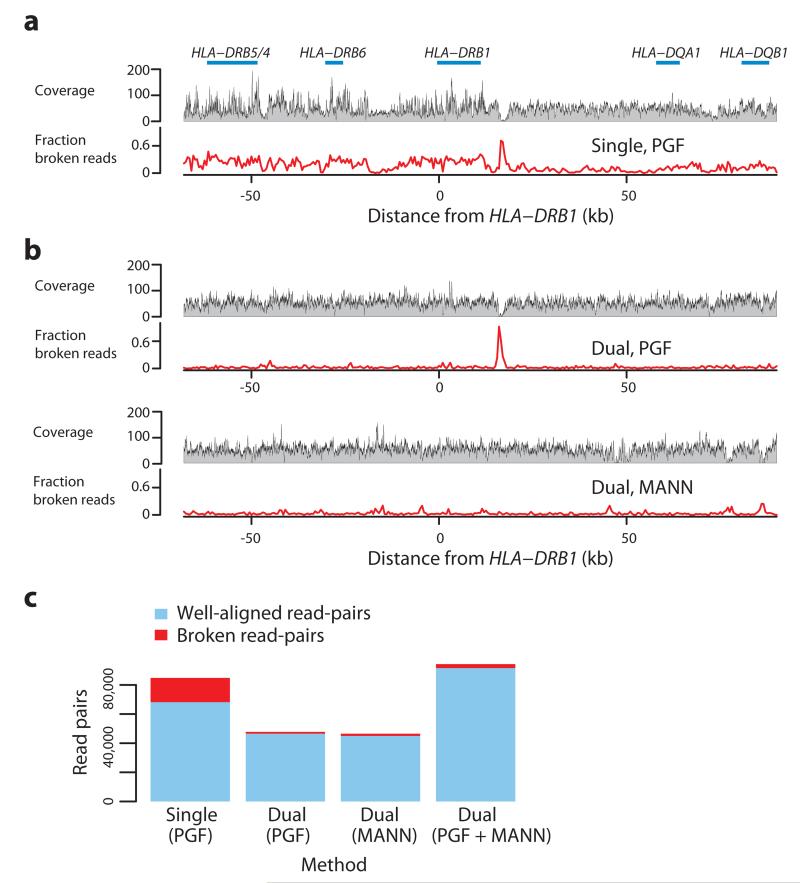
Figure 1. Read-mapping in the MHC Class II region.
a. Summary of read alignment to a single reference (GRCh37 without alternative loci, containing the ‘PGF’ haplotype in the xMHC) for a single sample (CS1) in the MHC Class II region (around HLA-DRB1) showing coverage (grey profile) and the proportion of ‘broken’ read-pairs (red line; defined as mapping to different chromosomes, incompatible strands, or implausible insert size). b The same metrics as for part a, where mapping has been performed to a reference augmented with the MANN (ALT_REF_LOCI_4, Genbank ID GL000253.1) haplotype (i.e. in addition to PGF), chosen because the combined classical HLA genotypes from PGF and MANN match those of the sample. c. Number of mapped intact (blue) and broken (red) read pairs for experiments underlying panels a and b (results from the latter split according to which haplotype reads map to, and a combined metric), demonstrating that the augmented reference results in many more well-mapped and many fewer broken read-pairs.