Significance
In the ParABS (partition system of ParA, ParB, and parS) bacterial chromosome partitioning system, ParB (partition protein B) spreads along the chromosomal DNA and a high-order complex is required for this function. Although ParB spreading has been studied, the detailed mechanism is still unclear. Herein, we report the crystal structure of the C-terminal domain truncated protein (Ct-HpSpo0J)-parS complex and reveal the structural basis for ParB spreading and chromosomal DNA condensation. Our structural and biochemical data show that ParB forms a high-order nucleoprotein: the ParB-parS complex. From these results, we propose how ParB possibly associates with other proteins through its protruded N-terminal domains, how ParB spreads along the chromosomal DNA by parS binding, and how ParB bridges DNA to compact and condense the chromosome during chromosome partitioning.
Keywords: arginine patch, chromosome segregation, ParABS, ParB spreading, parS
Abstract
Spo0J (stage 0 sporulation protein J, a member of the ParB superfamily) is an essential component of the ParABS (partition system of ParA, ParB, and parS)-related bacterial chromosome segregation system. ParB (partition protein B) and its regulatory protein, ParA, act cooperatively through parS (partition S) DNA to facilitate chromosome segregation. ParB binds to chromosomal DNA at specific parS sites as well as the neighboring nonspecific DNA sites. Various ParB molecules can associate together and spread along the chromosomal DNA. ParB oligomer and parS DNA interact together to form a high-order nucleoprotein that is required for the loading of the structural maintenance of chromosomes proteins onto the chromosome for chromosomal DNA condensation. In this report, we characterized the binding of parS and Spo0J from Helicobacter pylori (HpSpo0J) and solved the crystal structure of the C-terminal domain truncated protein (Ct-HpSpo0J)-parS complex. Ct-HpSpo0J folds into an elongated structure that includes a flexible N-terminal domain for protein–protein interaction and a conserved DNA-binding domain for parS binding. Two Ct-HpSpo0J molecules bind with one parS. Ct-HpSpo0J interacts vertically and horizontally with its neighbors through the N-terminal domain to form an oligomer. These adjacent and transverse interactions are accomplished via a highly conserved arginine patch: RRLR. These interactions might be needed for molecular assembly of a high-order nucleoprotein complex and for ParB spreading. A structural model for ParB spreading and chromosomal DNA condensation that lead to chromosome segregation is proposed.
The integrity of chromosomes and plasmids relies on precise DNA replication and segregation (1, 2). The initiation of DNA replication has to synchronize with the cell cycle to ensure precise chromosome segregation (3). In bacteria, the chromosome-encoded plasmid-partitioning system (Par) (4) and the structural maintenance of chromosomes (SMC) condensation complex (5) are two highly conserved systems associate with chromosome segregation and organization. SMC contributes to the overall stability and organization of genome (6–8). The partition system denoted ParABS is comprised of two proteins (ParA and ParB) and a centromere-like DNA element (parS) (9). ParB binds specifically to parS to form a complex. After binding ATP, ParA can interact with the ParB–parS complex to form a nucleoid–adaptor complex. ParB promotes the ATP hydrolysis activity of the complex to separate the chromosomes (9–13).
In the bacterial chromosomal ParABS system, ParB has two functions: one is to regulate chromosome replication and sporulation (8, 12, 14) and the other is to participate in chromosome segregation (5, 15–17). ParB spreads along the chromosomal DNA by binding at specific parS and nonspecific DNA sites to form a high-order partition complex (18–20). This partition complex is required for the loading of SMC onto the chromosomal DNA (5). In addition, the N-terminal domain of ParB can interact with ParA and stimulate its ATPase activity (21). This nucleoid–adaptor complex, ParA–ParB–parS is used to drive chromosome segregation (22, 23). However, the detailed mechanism for this process is still unclear.
Members of the ParB superfamily share similar functional domains: an N-terminal domain for protein–protein interactions, a central DNA-binding domain for parS binding, and a C-terminal domain for ParB dimerization (24). Two conserved N-terminal domain residues, Lys3 and Lys7, in the ParB from Bacillus subtilis (BsSpo0J), have been shown to interact with its regulatory protein BsSoj, a member of the ParA superfamily (3). The loss-of-function BsSpo0J R80A mutant was originally discovered by Autret et al. (25) and reportedly has disrupted focus formation by fluorescence microscopy. More recently, Graham et al. (20) showed that BsSpo0J bridges chromosomal DNA using single-molecule experiments. However, its R79A, R80A, and R82A mutants could not spread in vivo and did not bridge DNA in vitro. These highly conserved arginine residues were defined as an arginine patch (20). Furthermore, Broedersz et al. (26) studied the condensation and localization of ParB by computational simulation.
The crystal structures of ParB superfamily proteins have been reported for a DNA-free form of TtSpo0J (from Thermus thermophilus, containing the N-terminal and the DNA-binding domains) (10) and three complexes: the RP4–KorB-OB complex (from plasmid RP4, containing the DNA-binding domain) (27), the P1 ParB–parS complex (from Enterobacteria phage P1, containing the DNA-binding and the C-terminal domains) (28), and the F-SopB–sopC complex (from plasmid F, containing the DNA-binding domain) (29).
The Helicobacter pylori ParABS system consists of HpSoj (ParA), HpSpo0J (ParB), and parS DNA (30, 31). Herein, we report the crystal structure of a C-terminal domain truncated HpSpo0J (Ct-HpSpo0J)–parS complex. The N-terminal and the DNA-binding domains are present on Ct-HpSpo0J. The structural details of the complex in combination with results from EMSAs, fluorescence anisotropy assay, and small angle X-ray scattering (SAXS) allow us to propose a model for ParB spreading as it relates to chromosome segregation.
Results
HpSpo0J and parS Binding.
Full-length HpSpo0J (residues 1–290) and the C-terminal domain truncated Ct-HpSpo0J (residues 1–240) were expressed in and isolated from Escherichia coli. Size exclusion chromatography in conjunction with multiangle light scattering (SEC-MALS) measurements indicate that HpSpo0J (62.0 kDa) is dimeric and Ct-HpSpo0J is monomeric (29.7 kDa) in solution (Fig. S1A). On adding the parS DNA (16.1 kDa), the HpSpo0J–parS complex (73.5 kDa) was observed with a binding stoichiometry of two HpSpo0J to one parS (Fig. S1A).
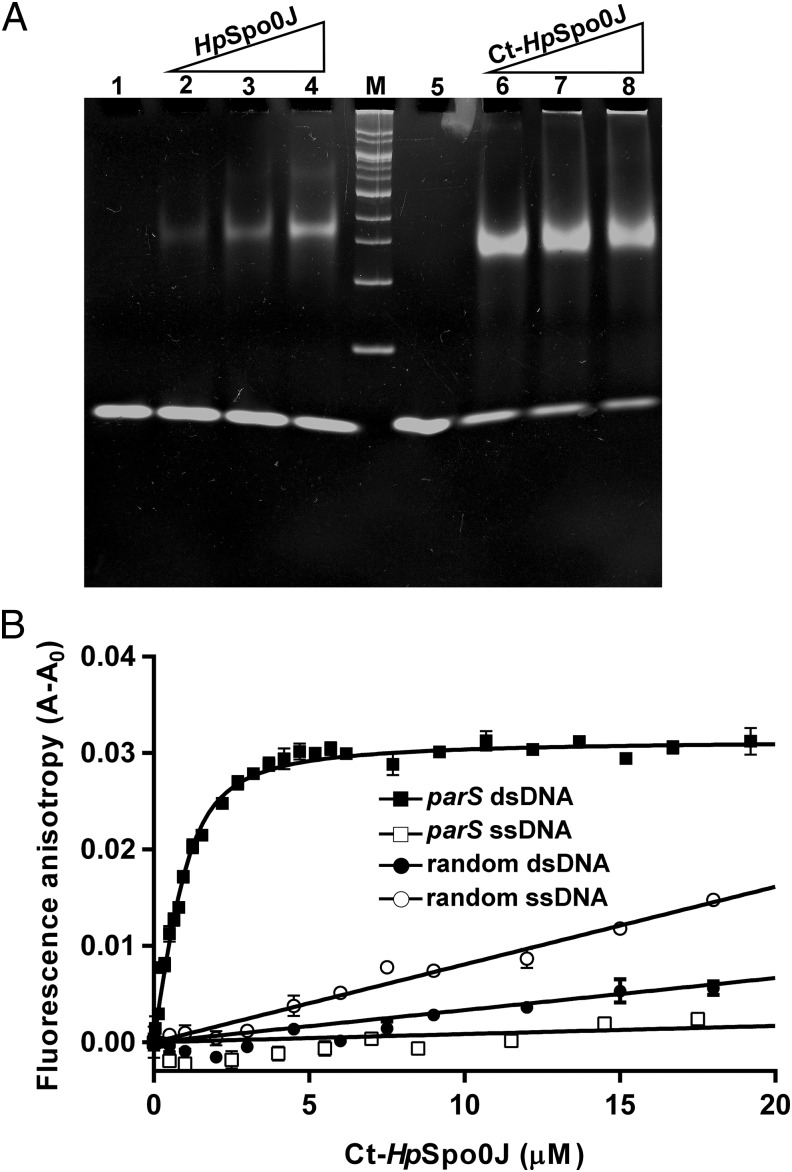
ParS binding by HpSpo0J or Ct-HpSpo0J was examined further with EMSA (Fig. 1A). HpSpo0J and Ct-HpSpo0J were complexed with parS at a protein to DNA ratio of 2:1 or greater. Furthermore, Ct-HpSpo0J has a higher affinity for parS than HpSpo0J. However, this observation is not conclusive because HpSpo0J tends to aggregate and precipitate. In addition, Ct-HpSpo0J binds specifically to double-stranded parS. It does not bind single-stranded parS or single- or double-stranded random DNA as shown by fluorescence anisotropy measurements (Fig. 1B). The dissociation constant of the Ct-HpSpo0J–parS complex is 0.31 ± 0.06 μM, similar to that of the P1 ParB–parS complex (32).
Fig. 1.
Ct-HpSpo0J–parS and HpSpo0J–parS complexes. (A) EMSAs. Various molar ratios (2, 4, 8) of HpSpo0J (lanes 2–4) and Ct-HpSpo0J (lanes 6–8) were mixed with parS (41 pmol) before electrophoresis. DNA markers (M: 100–2,000 bp) are shown. Lanes 1 and 5 contain only parS. (B) Fluorescence anisotropy. Various 5′-fluoresceine-labeled double- (■) and single-strand (□) parS and double- (●) and single-stranded (○) random DNAs (500 nM) were titrated with Ct-HpSpo0J (0–20 μM).
The Ct-HpSpo0J-parS Crystal Structure.
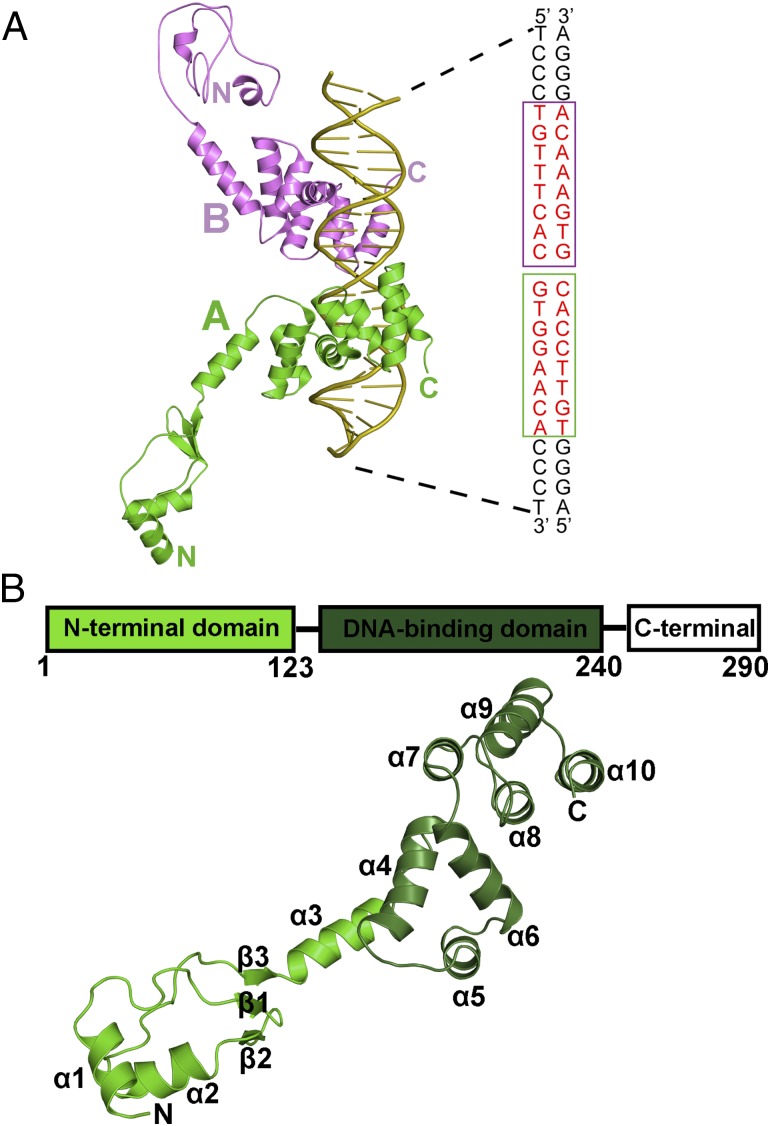
Ct-HpSpo0J was crystallized in the presence of parS (24 bp) at a ratio of 4:1, using PEG 8000 as the precipitant. The structure was determined at a resolution of 3.1 Å by the selenium multiwavelength anomalous dispersion method (33). The overall structure of the Ct-HpSpo0J–parS complex is shown in Fig. 2. Two Ct-HpSpo0J molecules interact with one parS. Each protein molecule interacts with half of the parS site and array on opposite sides of parS (Fig. 2A). Ct-HpSpo0J contains only the N-terminal (α1–3 and β1–3) and the DNA-binding (α4–10) domains. It folds into an elongated structure with a dimension of 78 Å (Fig. 2B). Ct-HpSpo0J binds to parS with the α5 and the α6 helices, forming a typical helix-turn-helix (HTH) binding motif. Arg159, Asn164, Lys190, Arg215, and Glu218 interact directly with the parS bases via hydrogen bonds. Residues Arg159 and Asn164 form specific interactions with the parS major groove. These two residues are conserved among most members of the ParB superfamily, with the exception of the P1 ParB, which has Gln179 and Arg184 at the corresponding positions (28). The Lys190 of Ct-HpSpo0J interacts with parS in a manner similar to that of the Arg219 of F-SopB (29, 34). However, interactions corresponding to those of Arg215 and Glu218 have not been reported for other systems. Although HpSpo0J shares the typical DNA-binding domain structure with P1 ParB, RP4-KorB, and F-SopB, several differences in their protein–DNA interactions exist. In addition, Gln148, Lys157, Ser158, Arg159, Arg167, Gly187, Val214, Arg222, and Arg225 on Ct-HpSpo0J interact with the phosphate backbone of the parS DNA. Numerous hydrophobic interactions between Ct-HpSpo0J and parS can also be observed. The detailed interactions between Ct-HpSpo0J and parS are listed in Table S1. In summary, the overall DNA-binding domain fold is broadly conserved with those of previously reported structures (27–29). However, certain unique interactions are present and might account for variations in parS recognition in different species.
Fig. 2.
The structure of the Ct-HpSpo0J-parS complex. (A) The Ct-HpSpo0J–parS complex. Two Ct-HpSpo0J molecules and one 24-bp parS DNA are shown. The two binding sites on a parS (16 bp, red), one each for the HpSpo0J A (green) and B (magenta) molecules, are boxed. (B) Ct-HpSpo0J monomer. A ribbon diagram of Ct-HpSpo0J is shown. The molecule contains 10 α helices (α1–10) and three β strands (β1–3). The N-terminal domain (green) and the DNA-binding domain (darker green) are shown.
Oligomerization Promoted by Adjacent and Transverse Interactions.
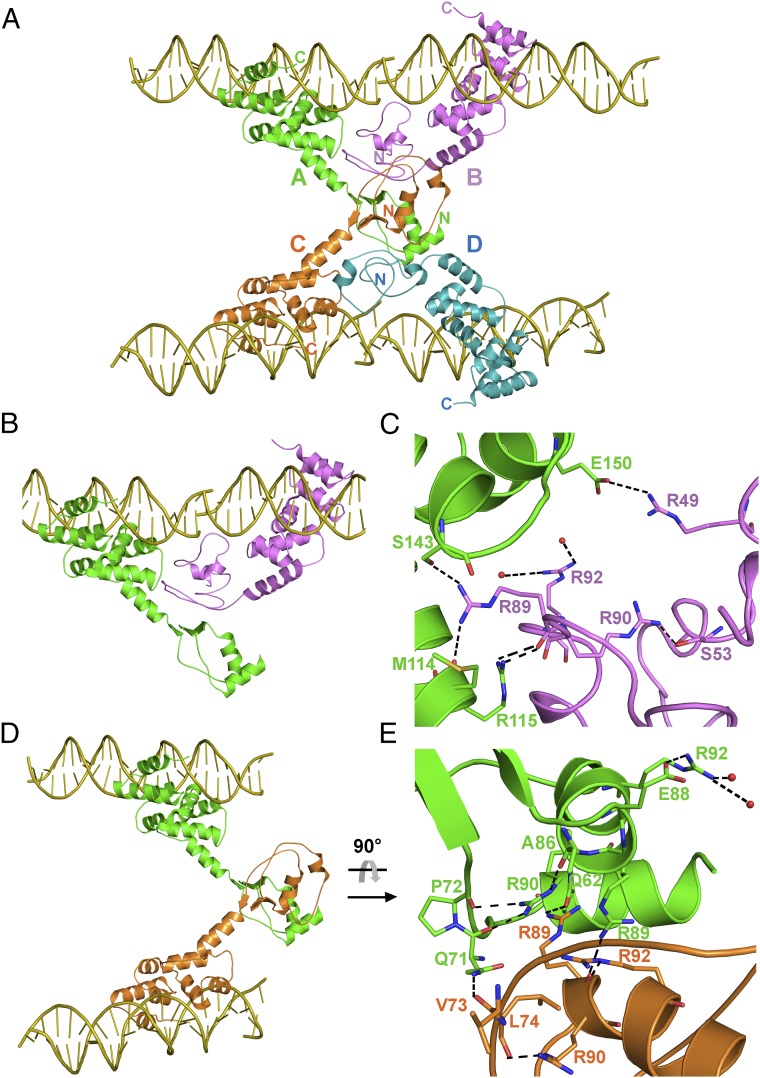
The oligomeric structure of the Ct-HpSpo0J–parS complex is formed by four Ct-HpSpo0J molecules (chains A, B, C, and D) binding with four parS molecules (Fig. 3A). The parS binding domain of the Ct-HpSpo0J–parS complex is preserved in the oligomer and has an overall structure similar to those of other ParB superfamily proteins (Fig. S2). However, their N-terminal domains are oriented differently. The N-terminal domain of the monomers participates in various protein–protein interactions when HpSpo0J exists in an oligomeric form. This oligomer might mimic a high-order complex in the ParABS system to promote ParB spreading. A highly cooperative molecular assembly is needed for ParB spreading, and the nucleoprotein complexes are essential in the partition system (18–20).
Fig. 3.
The oligomeric Ct-HpSpo0J–parS complex. (A) The Ct-HpSpo0J–parS complex. Within the crystal, four Ct-HpSpo0J molecules (A/green, B/magenta, C/orange, and D/cyan) and four parS DNAs (yellow) form as a tetramer. Each Spo0J monomer interacts with a parS half-site and only one of the two HpSpo0J molecules on each parS site is shown. (B) The AB dimer is formed by adjacent interactions. (C) Arginine patch, 89RRLR92, interactions at AB dimer. (D) The AC dimer is formed by transverse interactions. (E) Arginine patch, 89RRLR92, interactions at AC dimer.
The flexible N-terminal domain (α1–3 and β1–3) of the Ct-HpSpo0J is exposed and interacts to form AB, AC, and CD dimers, with buried interfaces of 753, 602, and 824 Å2, respectively. For the AB dimer, the B molecule interacts with the A molecule, by β1-β2, loop β2-α2, and α2. For the AC dimer, the C molecule interacts with the A molecule, by loop α1-β1, β1, loop β2-α2, and α2 (Fig. S3A). In both dimers, the B and the C molecules interact with the A molecule with different regions, except loop β2-α2 (residue Ala86) and α2 (residue Arg89) (Fig. S3A). For the AB and the AC dimers, the A molecule interacts with the B and the C molecules at distinct regions, using the linker helix (α3) to approach the B molecule and the two N-terminal end helices (α1 and α2) to contact the C molecule (Fig. S3B). The dimeric interactions of CD are similar to those of the AB dimer (Table S2). The B and the D molecules do not interact and appear to provide only adjacent interactions needed for the AB and the CD dimer formation but not the transverse interaction. Possibly, the N-terminal domains of the B and the D molecules might interact with other proteins, e.g., ParA/Soj. In summary, these dimer interfaces appears to be mainly promoted by the residues from α1–3 and β1–2 of the N-terminal domain. Residues that are interacting at the dimeric interfaces are listed in Table S2.
The N-terminal domain of ParB has been suggested to be essential for ParB spreading and DNA condensation during chromosome segregation (20, 35). Yamaichi et al. (4) identified two conserved motifs, boxes I and II, in the N-terminal domain of BsSpo0J. A mutagenesis study by Kusiak et al. (35) indicated that residues in the box I and the box II regions of P. aeruginosa ParB (PaParB) were involved in dimer formation. They have isolated a ParB oligomer by chemical crosslinking. The highly conserved residues Gly71/Arg94/Ala97 in PaParB were involved in a higher-order complex formation. Gly68/Arg90/Ala93 are the corresponding residues in HpSpo0J. Our finding that Arg90 involved in protein–protein interaction is consistent with Kusiak's results. However, the interactions for Gly68 and Ala93 cannot be found. Conserved residues in box I and II that participate in the oligomerization and play an important role in the higher-order complex formation are listed in Fig. S4 and Table S2.
A highly conserved “arginine patch” with the sequence “RRXR” is found in box II of ParB. Mutations of these arginines do not impair the protein binding to parS (20, 35). In the N-terminal domain of HpSpo0J, the corresponding box I (residues 63–76) is located between α1 and β1, and box II (residues 82–93) is located between β2 and α2 (Fig. 3 and Fig. S4). In the Ct-HpSpo0J–parS complex, residues from these two conserved boxes participate in oligomer formation but not in the parS binding (Fig. S4 and Table S2). The arginine patch in HpSpo0J, 89RRLR92 positioned at α2 faces a neighboring molecule, and the three arginines, Arg89/Arg90/Arg92, are involved in interchain interactions (Fig. 3).
In the Ct-HpSpo0J–parS complex, molecules A and B form a cross-wise dimer using adjacent molecular interactions via N-terminal domain residues (Fig. 3 A and B). Residues of α3 and α4 in chain A interact with Arg89 of chain B. Several hydrogen bonds are formed between Met114/Arg115 (α3) and Ser143 (α4) of chain A and Arg89 of chain B (Fig. 3C and Table S2). In addition, Glu150 (α5) of chain A forms a salt bridge with the conserved Arg49 of chain B. These hydrophilic interactions might be essential for AB dimer formation. In addition, the AB dimer formation involves hydrophobic interactions (Table S2) including mainly box II residues. The mutagenesis of the BsSpo0J Gly77, which is conserved in all ParB sequences except that of P1 ParB, indicated that the mutation (G77S) does not prevent ParB bridging in the spreading process (20). The corresponding residue in HpSpo0J is Gly87 which is involved in the dimer interaction in the Ct-HpSpo0J–parS complex.
Chains A and C form a bridging dimer via transverse molecular interactions of residues in their N-terminal domains (Fig. 3 A and D). For instance, a hydrogen bond forms between the side chain and main chain of Arg89 in chains A and C, respectively. In addition, Gln62 (α1) of chain A interacts with the side chain of Arg89 of chain C, and the chain A Gln71 (loop α1-β1) interacts with the chain C Val73 (Fig. 3E and Table S2). Hydrophobic interactions involving mainly box I residues (Table S2) also contribute to AC dimer formation.
In the Ct-HpSpo0J–parS complex, the molecular interactions between a cross-wise AB dimer are more extensive than those of the bridging AC dimer. In particular, Arg89 in the arginine patch participates in the dimer formation, whereas Arg90 and Arg92 might maintain the stability of molecule. Arg90 forms several hydrogen bonds with Gln71/Pro72/Ala86, which may stabilize loops α1-β1 and β2-α2. Arg92 forms hydrophilic interactions with Glu88 and water molecules, which may stabilize α2 (Fig. 3 C and E). However, a mutagenesis study is needed to confirm these speculations.
Structural Comparisons.
ParB has a central parS binding domain that connects to its N and C termini with flexible linkers (linkers 1 and 2), which might affect the orientations of the domains (30). In HpSpo0J, these two linkers encompass residues 110EQE112 and 241QTL243 (Figs. S4 and S5).
In the absence of DNA, TtSpo0J has a bent N-terminal domain that folds a tight dimer in a closed conformation to stabilize the overall structure (10). The DNA-binding domains of TtSpo0J and HpSpo0J can be well superimposed, and they share a similar HTH motif (α4–6; Fig. S5A). Although the N-terminal domains of both proteins have a similar fold, there is a distance of 64 Å between the Cα atoms of the Asp55 of Ct-HpSpo0J and the Glu45 of TtSpo0J. The N-terminal domain of the Ct-HpSpo0J–parS complex adopts an open conformation, suggesting that parS binding might induce a conformational transition. This conformational change might be needed for the assembly of the high-order ParB–parS complex. In contrast, Leonard et al. (10) proposed that parS binding might cause the dissociation of the C-terminal domain into an open conformation and thus available for oligomerization. However, the structure of TtSpo0J was determined in the absence of DNA and the C-terminal domain. Based on our Ct-HpSpo0J-parS complex structure, the N-terminal domain becomes an open conformation upon parS binding and is involved in protein–protein interaction.
Although their sequence homology is low, the Ct-HpSpo0J–parS and the P1 ParB–parS (1ZX4) (28) complexes have similar DNA-binding domains (α4–6 in Ct-HpSpo0J and α1–3 in P1 ParB) with an RMSD of 0.9 Å in Cα atoms (Fig. S5 B and C). The N-terminal domain of HpSpo0J and the C-terminal domain of P1 ParB are pointed away from their DNA-binding domains and protruded outward (Fig. S5B). The corresponding helices α7 and α10 of P1 ParB and Ct-HpSpo0J, respectively, are oriented in opposite directions (Fig. S5C). Consequently, α10 of the Ct-HpSpo0J–parS complex is involved in parS binding, whereas the α7 of the P1 ParB–parS complex participates in C-terminal domain dimerization. These differences might because of the P1 ParB–parS complex include the C-terminal domain and linker 2 that would trigger the C-terminal domain dimerization and modify the parS-bound state.
A full-length ParB structure is not currently available. From structural predications and comparisons of P1 ParB [Protein Data Bank (PDB) ID code 1ZX4], TtSpo0J (PDB ID code 1VZ0), and Ct-HpSpo0J (this study), they might share a similar structural topology (Fig. S5D). Therefore, a structural model of the full-length Spo0J can be built by aligning the DNA-binding domain of Ct-HpSpo0J and P1 ParB complexes for which the N- and C-terminal domains can be constructed, respectively. This structural model has an omega shape (Fig. S5 B and C). The N and C termini of this model appear to be exposed and could therefore orient in various directions.
Full-Length HpSpo0J.
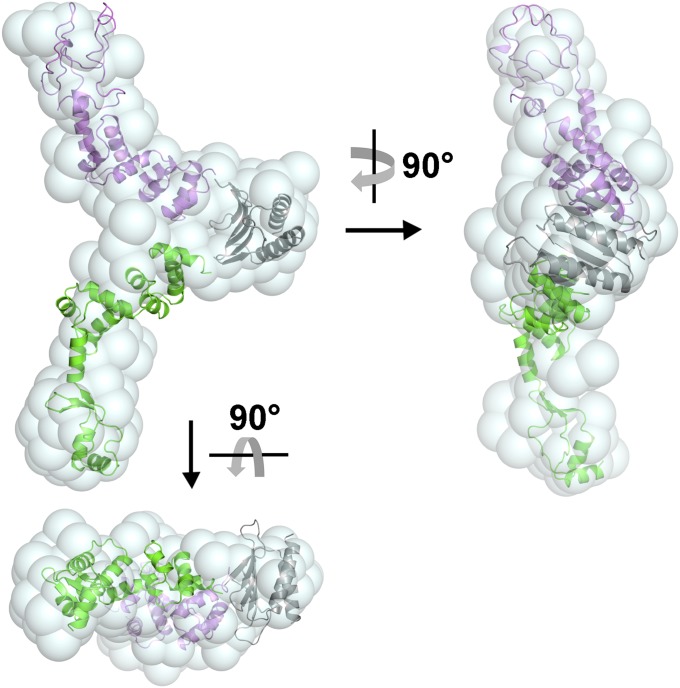
We used SAXS to further investigate the full-length HpSpo0J. The SAXS data was fitted by GNOM and the result are shown in Fig. S6 A and B, respectively. From the SAXS data, a nonglobular molecular envelope (Fig. 4) was generated using ab initio calculation. The overall structure of HpSpo0J was modeled by DAMMIN, using the structures of the Ct-HpSpo0J–parS complex (this study) and the P1 ParB–parS complex (28) as templates. Although the sequence homology of the C-terminal domains of HpSpo0J and P1 ParB is not high, they are predicated to share similar secondary structures (Fig. S5D). After fitting the C-terminal domain of P1 ParB into the nonglobular envelop, the N-terminal and DNA-binding domains of HpSpo0J were fit into the two arms of the molecular envelope (Fig. 4). Based on the SAXS data, the full-length HpSpo0J is a Y-shaped structure that might dimerize via its C-terminal domains, whereas its N-terminal domain is available for neighboring molecular interactions (Fig. 4). This suggestion is consistent with the SEC-MALS results (Fig. S1A), showing the full-length HpSpo0J forms a dimer in solution. However, we cannot exclude other possibilities because of the flexibility of the N-terminal domain and the low sequence homology of the C-terminal domain with that of P1 ParB.
Fig. 4.
A structural model of the full-length HpSpo0J. The superposition of the Y-shaped SAXS envelope and the structural model of the full-length HpSpo0J are shown. HpSpo0J is shown as a dimer. The N-terminal and DNA-binding domain of the two HpSpo0J molecules are colored separately in green and magenta, respectively. Their C-terminal domains are shown as a dimer and colored in gray.
Discussion
ParB binds specifically to parS sites and also associates with neighboring nonspecific DNA (14, 36). The N- and C-terminal domains of ParB may exhibit different functions during the course of chromosome segregation (10). The C-terminal domain serves primarily as a region for dimerization (10). The flexible N-terminal domain is responsible for molecular spreading with a neighboring ParB and interactions with ParA and SMC (12, 20). ParB spreading is essential for recruiting multiple and remote DNAs around the parS site (20). The higher-order partition complex needs to be formed, and the chromosomal DNA needs to be compacted and condensed for chromosome segregation.
In the Ct-HpSpo0J–parS complex, AB and AC dimer formation occurs via their N-terminal domains by various molecular interactions, structurally and functionally (Fig. 3). Adjacent interactions involve residues in chain A α3 and α4 to interact with those in chain B α2 to form the AB dimer. Alternatively, transverse interactions of the AC dimer require the residues in chain A α1 and α2 to interact with those in chain C α2. These adjacent and transverse molecular interactions involving the N-terminal domain might mimic how ParB spreads after binding parS. The highly conserved Arg89 in the arginine patch also provides important interactions for cross-wise and bridging dimers formation.
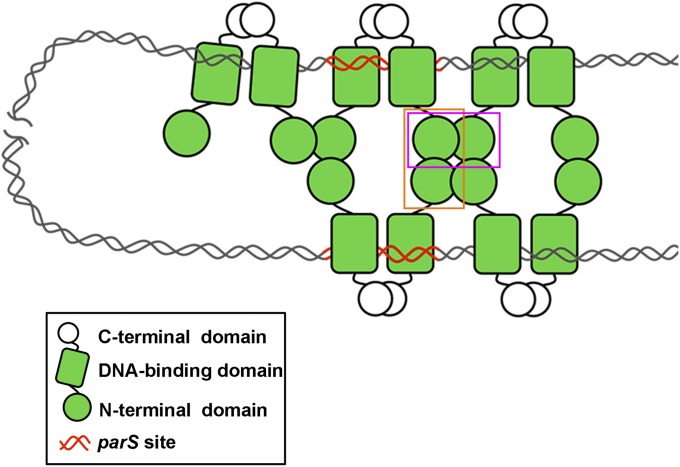
Accordingly, we propose a ParB spreading model (Fig. 5). In this model, all three domains of ParB participate cooperatively for molecular spreading. In this process, the DNA-binding domain binds to chromosome at a specific parS site. The C-terminal domain of ParB dimerizes with a neighbor to stabilize the ParB–parS complex. The flexible and protruded N-terminal domain provides multiple protein–protein interactions. The neighboring ParB dimers interact by adjacent interactions through the N-terminal domain and spread along the chromosomal DNA, horizontally. In addition, using transverse interactions via the N-terminal domain, the bridging ParB dimers recruit the distal chromosomal DNA vertically. These molecular interactions assemble the ParB molecules into a tetrameric-like oligomer. Taking in the specific and nonspecific DNA binding of ParB, a high-order nucleoprotein complex can be built and situated on the chromosome. ParB spreading takes place spatially and the remote chromosomal DNA can be engaged by looping and compacted. Subsequently, the auxiliary ATPase protein ParA and SMC protein could be recruited, and the chromosomal DNA is condensed. Finally, DNA segregation will occur.
Fig. 5.
ParB spreading model. The C-terminal dimerization (in white sphere), the DNA-binding (in green rectangle), and the flexible N-terminal domains (in green sphere) of ParB are shown. A ParB dimer is formed via the C-terminal dimerization domain. ParB binds chromosomal DNA at the specific parS site (in red) or nonspecific sites (in gray). Multiple ParB molecules spreading along the lengthy and looping chromosomal DNA through the N-terminal domain by adjacent interactions (boxed in magenta) horizontally and transverse interactions (boxed in orange) vertically. Consequently, a high-order ParB–DNA complex is formed.
Only a small number of Spo0J molecules are found spreading over many kilobases of DNA. This observation is not consistent with the quantity of Spo0J in cells (20). Graham et al. (20) suggested that molecular spreading involved DNA bridging and that nearest-neighbor interactions associate ∼20 Spo0J dimers with a parS. Broedersz et al. (26) suggested a ParB-spreading model that combines one-dimensional spreading with 3D bridging of ParB proteins on DNA for the formation of a coherent complex. These biochemical and in vivo results strongly support the physiological relevance of the HpSpo0J, as the arginine patch (R89, R90, and R92) was observed to form part of the oligomerization motif (Fig. 3). Our structural data and ParB spreading model are consistent with observations concerning the chromosome partition system. Based on the Ct-HpSpo0J–parS complex structure (Fig. 3), long-range DNA looping can be achieved via the cooperation of transverse and adjacent interactions to assemble a high-order ParB–parS complex that span many kilobases of DNA. The significant molecular interactions found in the Ct-HpSpo0J–parS complex provide a mechanism to mimic ParB spreading in the chromosome partition system.
Materials and Methods
EMSA.
The DNA binding activities of HpSpo0J and Ct-HpSpo0J to parS were assayed using an EMSA. Reaction mixtures contain 2.4, 4.7, and 9.4 μL of HpSpo0J (29 μM) or Ct-HpSpo0J (29 μM) and 0.4 μL parS (100 μM) in molar ratios of 2, 4, and 8 (protein: DNA), respectively (Fig. 1A). Each mixture was brought to a final volume of 15 μL of an elution buffer at 25 °C for 15 min. The samples were then resolved through 8% (wt/vol) polyacrylamide (19:1) gel in 1× Tris/Borate/EDTA (TBE) buffer at 80 V and 4 °C for 3 h.
Crystallization.
The Ct-HpSpo0J–parS complex crystal was grown using the hanging-drop vapor diffusion method. Ct-HpSpo0J (20 mg/mL) was mixed with parS at a molar ratio of 4:1 (protein: DNA) in the reaction buffer. The protein sample was mixed and then equilibrated against precipitant solution at 293 K. The Ct-HpSpo0J–parS complex crystals grown by precipitant solution [14–16% (vol/vol) PEG 8000 and 450–525 mM lithium sulfate] within 3–7 d and grew to dimensions of 0.3 × 0.1 × 0.1 mm.
Detailed descriptions of experiments are provided in SI Materials and Methods. In general, full-length HpSpo0J (residues 1–290) and Ct-HpSpo0J (residues 1–240) were expressed in E. coli strain SG13009 and purified by Ni-affinity chromatography (Fig. S1B). The Ct-HpSpo0J–parS complex was crystallized, and the structure was determined by the multiwavelength anomalous dispersion method (33). The structural refinement was carried out by CNS (37) and PHENIX (38). The statistics of X-ray diffraction data and structural refinements are summarized in Table S3. The dissociation constant (Kd) was determined by fluorescence anisotropy using Prism (GraphPad Software). The solution state of HpSpo0J and Ct-HpSpo0J were determined by SEC-MALS. The structural model of the full-length HpSpo0J in solution was determined by SAXS.
Supplementary Material
Acknowledgments
We thank Dr. C. D. Hsiao and Dr. M. F. Tam for helpful discussion and suggestions. We are grateful for the access to the synchrotron radiation beamlines 13B1 and 13C1 at the National Synchrotron Radiation Research Center (NSRRC) and the in-house X-ray facility in the Macromolecular X-ray Crystallographic Center of National Tsing Hua University. We thank Dr. S. T. Danny Hsu for assistance with the SEC-MALS experiment and Dr. U. S. Jeng for assistance with the SAXS experiment performed at the NSRRC 23A beamline. This work was supported by Ministry of Science and Technology, Taiwan Grants 101-2311-B-007-009-MY3 and 103-2627-M-007-008 (to Y.-J.S.), and National Tsing Hua University, Taiwan Grant 102N2787E1 (to Y.-J.S.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 4UMK).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1421927112/-/DCSupplemental.
References
- 1.Lin DC, Grossman AD. Identification and characterization of a bacterial chromosome partitioning site. Cell. 1998;92(5):675–685. doi: 10.1016/s0092-8674(00)81135-6. [DOI] [PubMed] [Google Scholar]
- 2.Kadoya R, Baek JH, Sarker A, Chattoraj DK. Participation of chromosome segregation protein ParAI of Vibrio cholerae in chromosome replication. J Bacteriol. 2011;193(7):1504–1514. doi: 10.1128/JB.01067-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Scholefield G, Whiting R, Errington J, Murray H. Spo0J regulates the oligomeric state of Soj to trigger its switch from an activator to an inhibitor of DNA replication initiation. Mol Microbiol. 2011;79(4):1089–1100. doi: 10.1111/j.1365-2958.2010.07507.x. [DOI] [PubMed] [Google Scholar]
- 4.Yamaichi Y, Niki H. Active segregation by the Bacillus subtilis partitioning system in Escherichia coli. Proc Natl Acad Sci USA. 2000;97(26):14656–14661. doi: 10.1073/pnas.97.26.14656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sullivan NL, Marquis KA, Rudner DZ. Recruitment of SMC by ParB-parS organizes the origin region and promotes efficient chromosome segregation. Cell. 2009;137(4):697–707. doi: 10.1016/j.cell.2009.04.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wu N, Yu H. The Smc complexes in DNA damage response. Cell Biosci. 2012;2:5–15. doi: 10.1186/2045-3701-2-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Losada A, Hirano T. Dynamic molecular linkers of the genome: The first decade of SMC proteins. Genes Dev. 2005;19(11):1269–1287. doi: 10.1101/gad.1320505. [DOI] [PubMed] [Google Scholar]
- 8.Gruber S, Errington J. Recruitment of condensin to replication origin regions by ParB/SpoOJ promotes chromosome segregation in B. subtilis. Cell. 2009;137(4):685–696. doi: 10.1016/j.cell.2009.02.035. [DOI] [PubMed] [Google Scholar]
- 9.Erdmann N, Petroff T, Funnell BE. Intracellular localization of P1 ParB protein depends on ParA and parS. Proc Natl Acad Sci USA. 1999;96(26):14905–14910. doi: 10.1073/pnas.96.26.14905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Leonard TA, Butler PJ, Löwe J. Structural analysis of the chromosome segregation protein Spo0J from Thermus thermophilus. Mol Microbiol. 2004;53(2):419–432. doi: 10.1111/j.1365-2958.2004.04133.x. [DOI] [PubMed] [Google Scholar]
- 11.Livny J, Yamaichi Y, Waldor MK. Distribution of centromere-like parS sites in bacteria: Insights from comparative genomics. J Bacteriol. 2007;189(23):8693–8703. doi: 10.1128/JB.01239-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Havey JC, Vecchiarelli AG, Funnell BE. ATP-regulated interactions between P1 ParA, ParB and non-specific DNA that are stabilized by the plasmid partition site, parS. Nucleic Acids Res. 2012;40(2):801–812. doi: 10.1093/nar/gkr747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang X, Montero Llopis P, Rudner DZ. Bacillus subtilis chromosome organization oscillates between two distinct patterns. Proc Natl Acad Sci USA. 2014;111(35):12877–12882. doi: 10.1073/pnas.1407461111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Breier AM, Grossman AD. Whole-genome analysis of the chromosome partitioning and sporulation protein Spo0J (ParB) reveals spreading and origin-distal sites on the Bacillus subtilis chromosome. Mol Microbiol. 2007;64(3):703–718. doi: 10.1111/j.1365-2958.2007.05690.x. [DOI] [PubMed] [Google Scholar]
- 15.Leonard TA, Butler PJ, Löwe J. Bacterial chromosome segregation: Structure and DNA binding of the Soj dimer—A conserved biological switch. EMBO J. 2005;24(2):270–282. doi: 10.1038/sj.emboj.7600530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang X, Tang OW, Riley EP, Rudner DZ. The SMC condensin complex is required for origin segregation in Bacillus subtilis. Curr Biol. 2014;24(3):287–292. doi: 10.1016/j.cub.2013.11.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kleine Borgmann LA, Hummel H, Ulbrich MH, Graumann PL. SMC condensation centers in Bacillus subtilis are dynamic structures. J Bacteriol. 2013;195(10):2136–2145. doi: 10.1128/JB.02097-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Funnell BE. How to build segregation complexes in bacteria: Use bridges. Genes Dev. 2014;28(11):1140–1142. doi: 10.1101/gad.244517.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kim SK, Wang JC. Gene silencing via protein-mediated subcellular localization of DNA. Proc Natl Acad Sci USA. 1999;96(15):8557–8561. doi: 10.1073/pnas.96.15.8557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Graham TGW, et al. ParB spreading requires DNA bridging. Genes Dev. 2014;28(11):1228–1238. doi: 10.1101/gad.242206.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Radnedge L, Youngren B, Davis M, Austin S. Probing the structure of complex macromolecular interactions by homolog specificity scanning: The P1 and P7 plasmid partition systems. EMBO J. 1998;17(20):6076–6085. doi: 10.1093/emboj/17.20.6076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Murray H, Errington J. Dynamic control of the DNA replication initiation protein DnaA by Soj/ParA. Cell. 2008;135(1):74–84. doi: 10.1016/j.cell.2008.07.044. [DOI] [PubMed] [Google Scholar]
- 23.Rodionov O, Yarmolinsky M. Plasmid partitioning and the spreading of P1 partition protein ParB. Mol Microbiol. 2004;52(4):1215–1223. doi: 10.1111/j.1365-2958.2004.04055.x. [DOI] [PubMed] [Google Scholar]
- 24.Schumacher MA. Structural biology of plasmid partition: Uncovering the molecular mechanisms of DNA segregation. Biochem J. 2008;412(1):1–18. doi: 10.1042/BJ20080359. [DOI] [PubMed] [Google Scholar]
- 25.Autret S, Nair R, Errington J. Genetic analysis of the chromosome segregation protein Spo0J of Bacillus subtilis: Evidence for separate domains involved in DNA binding and interactions with Soj protein. Mol Microbiol. 2001;41(3):743–755. doi: 10.1046/j.1365-2958.2001.02551.x. [DOI] [PubMed] [Google Scholar]
- 26.Broedersz CP, et al. Condensation and localization of the partitioning protein ParB on the bacterial chromosome. Proc Natl Acad Sci USA. 2014;111(24):8809–8814. doi: 10.1073/pnas.1402529111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Khare D, Ziegelin G, Lanka E, Heinemann U. Sequence-specific DNA binding determined by contacts outside the helix-turn-helix motif of the ParB homolog KorB. Nat Struct Mol Biol. 2004;11(7):656–663. doi: 10.1038/nsmb773. [DOI] [PubMed] [Google Scholar]
- 28.Schumacher MA, Funnell BE. Structures of ParB bound to DNA reveal mechanism of partition complex formation. Nature. 2005;438(7067):516–519. doi: 10.1038/nature04149. [DOI] [PubMed] [Google Scholar]
- 29.Schumacher MA, Piro KM, Xu W. Insight into F plasmid DNA segregation revealed by structures of SopB and SopB-DNA complexes. Nucleic Acids Res. 2010;38(13):4514–4526. doi: 10.1093/nar/gkq161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gerdes K, Møller-Jensen J, Bugge Jensen R. Plasmid and chromosome partitioning: Surprises from phylogeny. Mol Microbiol. 2000;37(3):455–466. doi: 10.1046/j.1365-2958.2000.01975.x. [DOI] [PubMed] [Google Scholar]
- 31.Lee MJ, Liu CH, Wang SY, Huang CT, Huang H. Characterization of the Soj/Spo0J chromosome segregation proteins and identification of putative parS sequences in Helicobacter pylori. Biochem Biophys Res Commun. 2006;342(3):744–750. doi: 10.1016/j.bbrc.2006.01.173. [DOI] [PubMed] [Google Scholar]
- 32.Vecchiarelli AG, Schumacher MA, Funnell BE. P1 partition complex assembly involves several modes of protein-DNA recognition. J Biol Chem. 2007;282(15):10944–10952. doi: 10.1074/jbc.M611250200. [DOI] [PubMed] [Google Scholar]
- 33.Hendrickson WA, Ogata CM. Phase determination from multiwavelength anomalous diffraction measurements. Methods Enzymol. 1997;276(Macromolecular Crystallography, Part A):494–523. doi: 10.1016/S0076-6879(97)76074-9. [DOI] [PubMed] [Google Scholar]
- 34.Sanchez A, Rech J, Gasc C, Bouet JY. Insight into centromere-binding properties of ParB proteins: A secondary binding motif is essential for bacterial genome maintenance. Nucleic Acids Res. 2013;41(5):3094–3103. doi: 10.1093/nar/gkt018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kusiak M, Gapczynska A, Plochocka D, Thomas CM, Jagura-Burdzy G. Binding and spreading of ParB on DNA determine its biological function in Pseudomonas aeruginosa. J Bacteriol. 2011;193(13):3342–3355. doi: 10.1128/JB.00328-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Murray H, Ferreira H, Errington J. The bacterial chromosome segregation protein Spo0J spreads along DNA from parS nucleation sites. Mol Microbiol. 2006;61(5):1352–1361. doi: 10.1111/j.1365-2958.2006.05316.x. [DOI] [PubMed] [Google Scholar]
- 37.Brünger AT, et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr D Biol Crystallogr. 1998;54(Pt 5):905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 38.Adams PD, et al. PHENIX: Building new software for automated crystallographic structure determination. Acta Crystallogr D Biol Crystallogr. 2002;58(Pt 11):1948–1954. doi: 10.1107/s0907444902016657. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.