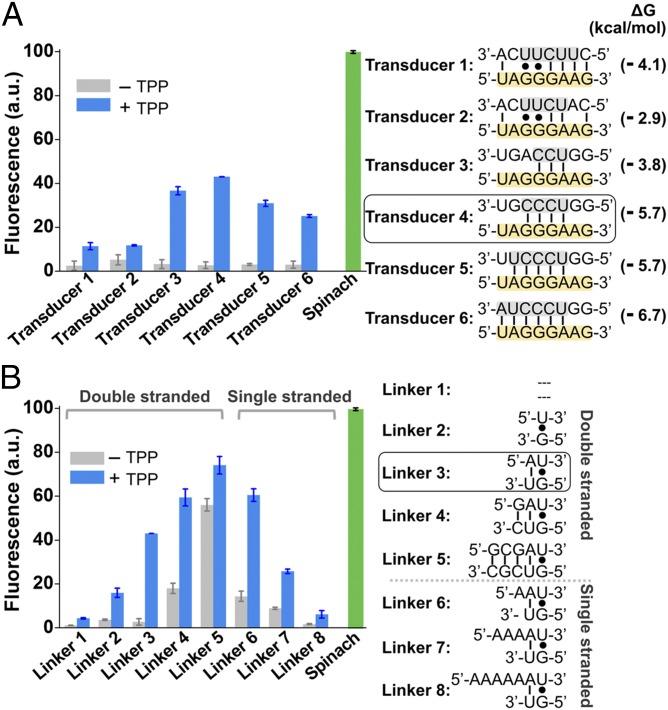
Fig. 2.
Optimization of Spinach riboswitch performance. (A) Optimization of transducer sequences. Six transducer sequences (transducers 1–6, gray) were tested for their ability to mediate TPP-induced fluorescence of the TPP Spinach riboswitch. In each case, the transducer sequence is similar to the transducer sequence found in the wild-type thiM TPP riboswitches. In each case, the switching sequence (yellow) in the aptamer domain is unchanged. The optimal sensor (transducer 4) contained a transducer sequence helix comprising only four Watson–Crick base pairs. The 5′-U base of this transducer sequence is positioned to form the U⋅A⋅U base triple that serves as the upper surface of the DFHBI-binding pocket. Shown are mean and SEM values of three independent replicates. The Gibbs free energy change (∆G) of hybridization events between each transducer sequence and the switching sequence was calculated using Mfold online software. (B) Optimization of linker sequences. Spinach was connected with the aptamer domain by different linker sequences. The linker position is highlighted in dark gray in Fig. 1B. The linker was one of eight double-stranded or single-stranded sequences. The double-stranded linkers of different lengths (linkers 1–5) extend the lower helix (H2) and rotate Spinach. The different single-stranded linkers (linkers 6–8) provide flexibility of relative domain positions that could facilitate helix formation. Shown are mean and SEM values of three independent replicates. The optimal linker (linker 3) displayed a 15.9-fold increase in fluorescence signal upon the addition of 100 µM TPP. a.u., arbitrary units.