Figure 1.
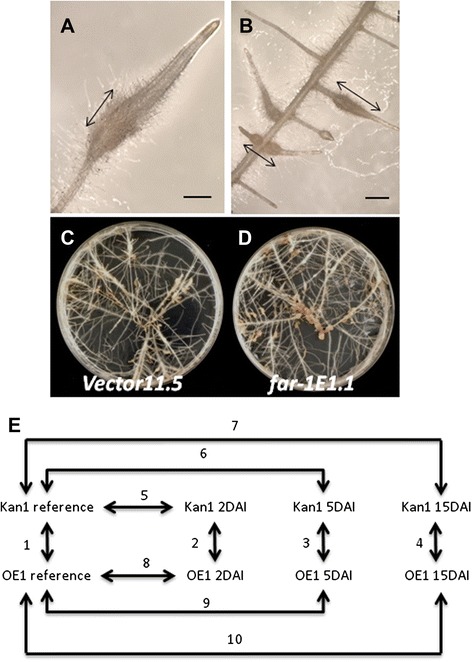
Plant material used for RNA-Seq analysis and the experimental design for complete tomato RNA-Seq profiling of tomato root lines carrying the kanamycin-resistance gene (Kan) or overexpressing mj-far-1 (OE) inoculated with Meloidogyne javanica . A representative 5 DAI gall used for RNA extraction on roots of the tomato control Kan (A) and OE line (B) following inoculation with Meloidogyne javanica. Arrows indicate segments collected for RNA extraction. Bars = 150 μm. Late infection stage of the tomato control Kan line (C) and OE line (D) as shown at 15 DAI. Note the increased size and density of galls on the OE vs. Kan roots. (E) Schematic representation of the experimental procedure. Four comparisons (1–4) between control Kan and OE lines were performed at each time point: 0, 2, 5, and 15 DAI. In addition, the reference tissue of each root line was directly compared with the transcript profile of the same root line at 2, 5, and 15 DAI: comparisons 5–7 for Kan and 8–10 for OE lines.
