Fig. 1. Expression cloning of vitamin D 25-hydroxylase cDNA.
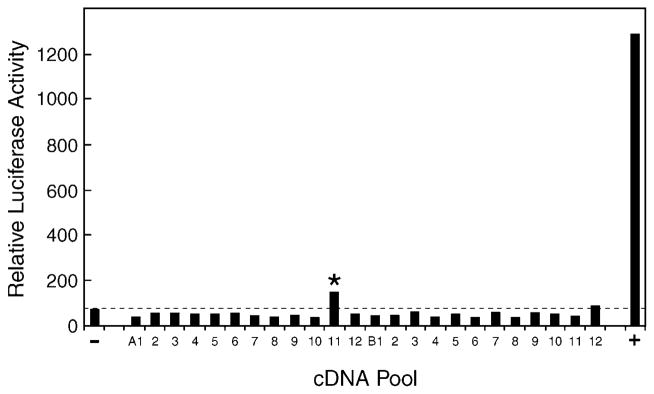
A cDNA library was constructed from hepatic mRNA purified from sterol 27-hydroxylase-deficient mice and divided into pools of ~100 cDNAs each. Plasmid DNA (75 ng) from individual pools was combined with 20 ng of a GAL4-vitamin D receptor expression plasmid, 20 ng of a GAL4-responsive luciferase reporter gene plasmid, 20 ng of a CMV-β-galactosidase plasmid (for normalization purposes), 5 ng of a mouse adrenodoxin expression plasmid (to boost the activity of mitochondrial P450s encoded by the cDNA pools), and 10 ng of the VA1 plasmid (to increase expression from the receptor and reporter gene plasmids; Ref. 30), and introduced via the FuGENE™ 6 reagent (Roche) into HEK 293 cells cultured in microtiter plates. 1α-Hydroxyvitamin D3 (10 nM) was added to the medium 8–10 h later. After an additional incubation of 16–20 h in an atmosphere of 91.2% air and 8.8% CO2, the cells were disrupted in the microtiter plate by the addition of 100 μl of Lysis Buffer, and aliquots of 20 and 40 μl were assayed by luminometry and spectroscopy, respectively, for luciferase and β-galactosidase enzyme activity. Relative luciferase activity was calculated for each well by dividing luciferase enzyme activity by the amount of β-galactosidase activity per unit of time. One pool of cDNAs (A11, marked by asterisk) produced a modest activation of the vitamin D receptor-reporter system compared with that obtained with the pCMV vector alone (−) and a positive control (+) consisting of a plasmid encoding the mouse CYP27A1 vitamin D 25-hydroxylase.
