FIGURE 1.
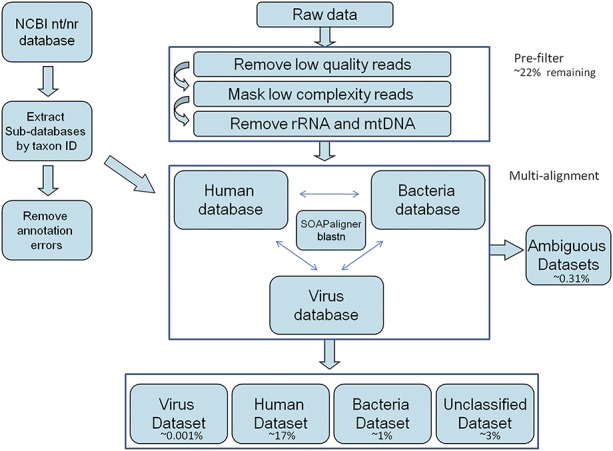
Bioinformatic pipeline for metagenomics. Reads were prefiltered and removed if they were low quality/complexity (∼7%), ribosomal RNA (∼69%), or mitochondrial RNA (∼6%). The remaining reads were assigned as human (∼17%) including HERV (∼0.006%), bacterial (∼1%), viral (∼0.001%), ambiguous (∼0.31%), or unclassified (∼3%) based on SOAPaligner or blastn.
