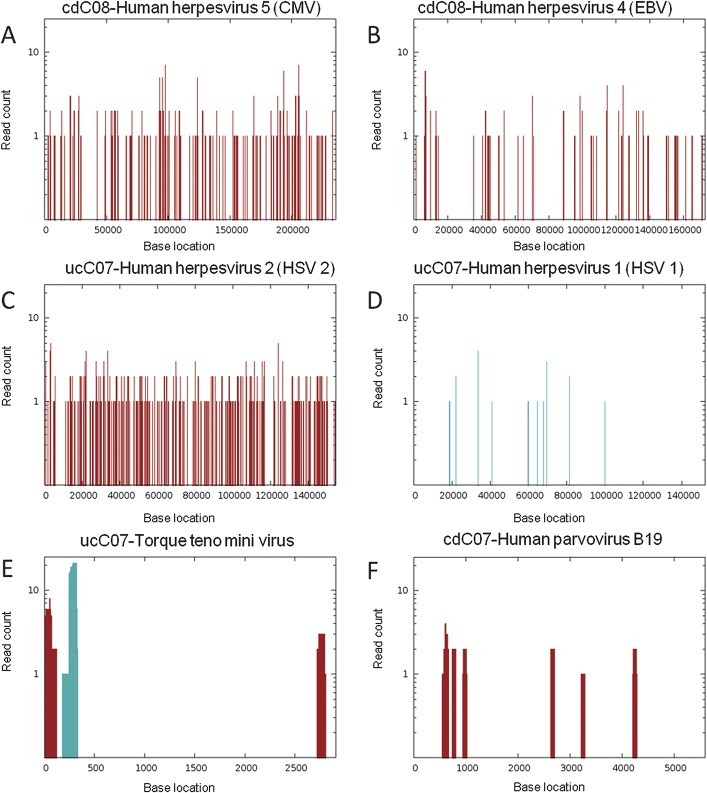
FIGURE 2.
Viral reads from individual libraries were aligned against reference viral genomes based on blastn to reference genomes, where red bars indicate number of reads that align only to 1 genome, whereas blue bars represent number of reads that align to multiple related genomes. Evidence for combined herpesviridae infection was observed in sample cdC08 with (A) 286 reads matching approximately 19 kB of CMV genome and (B) 195 reads covering 12 kB of EBV genome (CMV, gi 155573622; EBV, gi 139424478). In sample ucC07, 434 reads matched HSV with a coverage of (C) 25,944 bp for the HSV-2 genome and (D) 14081 bp for the HSV-1 genome, suggesting the presence of HSV-2 (HSV-2, gi 9629267; HSV-1, gi 9629378). (E) Torque teno mini virus was also detected in sample ucC07 (gi 295441877). (F), Human parvovirus B19 was detected in sample cdC07 (gi 9632996).