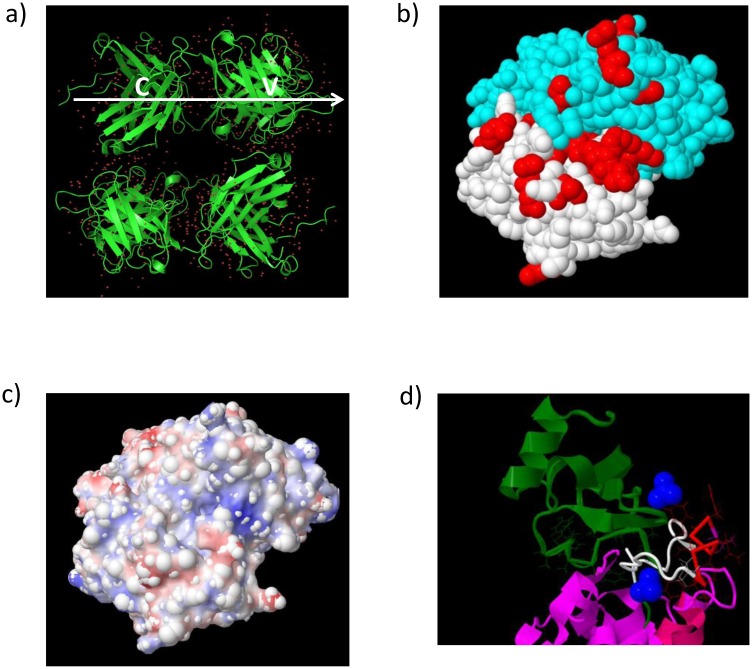
Fig 9. The crystal structure of RoAb13.
A, The full structure of the RoAb13 Fab fragment showing two dimers in the asymmetric unit plus waters showing the position of the constant (C) and variable (V) domains for the upper molecule. B, A space filling model of RoAb13 structure. The molecule is oriented with the membrane distal surface (the antigen binding surface) upwards (the model shown in panel a is rotated so the white arrow shown is pointing upwards perpendicular to the page). The position of the amino acids which differ between germline and RoAb13 are shown in red. C, The electrostatic antigen binding surface of RoAb13. The orientation is the same as shown in b). Negatively charged areas are shown in red, while positively charged areas are shown in blue. D, The structure of CCR5 bound to its ligand, CCL5. The model is based on the pdb file (ccl5_ccr5.10.pdb) containing the coordinates from the optimum MD simulation described in [39], displayed and edited in Jmol. CCL5 is shown in green. The N-terminal domain of CCR5 is shown in white, except for the linear epitope 8–16, which is shown in red. The two tyrosine sulphate groups are shown in blue (spacefill).