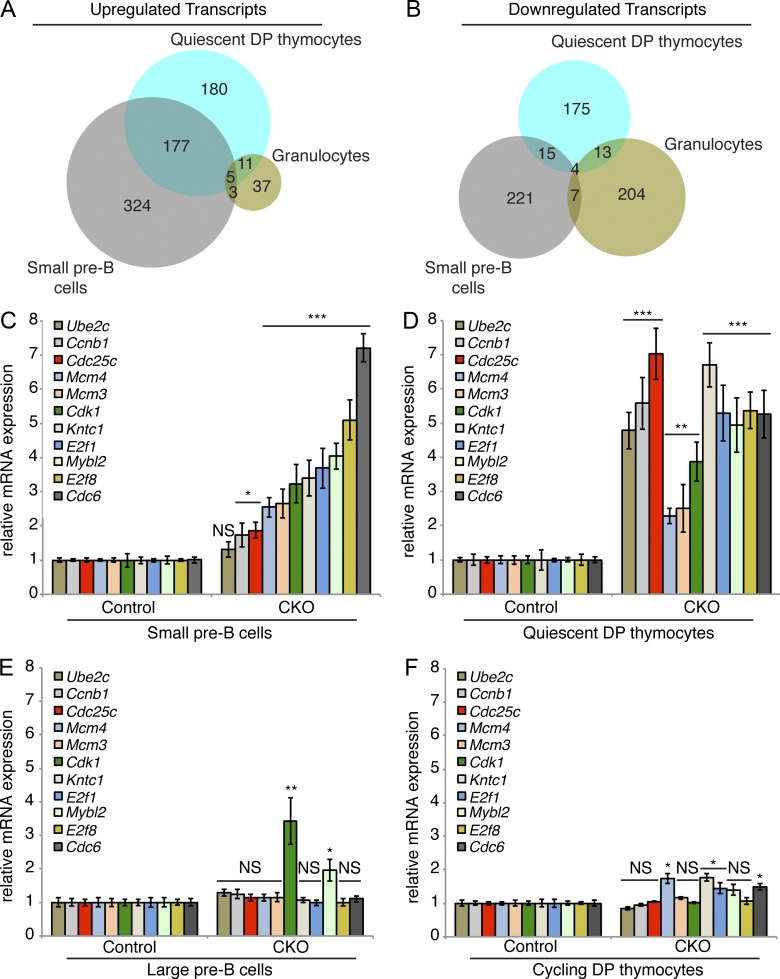
Figure 6.
Dyrk1a-deficient quiescent DP thymocytes and small pre–B cells fail to repress E2F target gene transcription. (A and B) Up- and down-regulated transcripts were identified by RNA-sequencing of FACS-purified quiescent DP thymocytes (CD4+/CD8+/DNA content2N/Pyronin Ylow), cycling DP thymocytes (CD4+/CD8+/DNA content>2N/Pyronin Yhigh), small pre–B cells (IgM−/B220+/CD43−/FSClow), large pre–B cells (IgM−/B220+/CD43low/FSChigh), and granulocytes (Gr-1high/Mac-1/CD11bhigh) from Dyrk1af/f Mx1-Cre− (Control) and Dyrk1af/f Mx1-Cre+ (CKO) mice. Venn diagrams depict shared up-regulated (A) and down-regulated (B) transcripts identified by RNA-sequencing in the three quiescent cell populations. (C–F) mRNA expression of E2F target genes was assessed by qRT-PCR in FACS-purified small pre–B cells (C), quiescent DP thymocytes (D), large pre–B cells (E), and cycling DP thymocytes (F) from Control and CKO mice 2 wk after pI:pC treatment. Transcript levels were normalized to Actb expression. PCRs were performed using 3 independently sorted pairs of samples (each with 1 mouse per genotype) for pre–B cells, and pooled samples from 3 mice per genotype for thymocytes. Error bars depict SD of triplicate wells for representative samples. *, P < 0.05; **, P < 0.01; ***, P < 0.001; NS, not significant (CKO vs. Control).