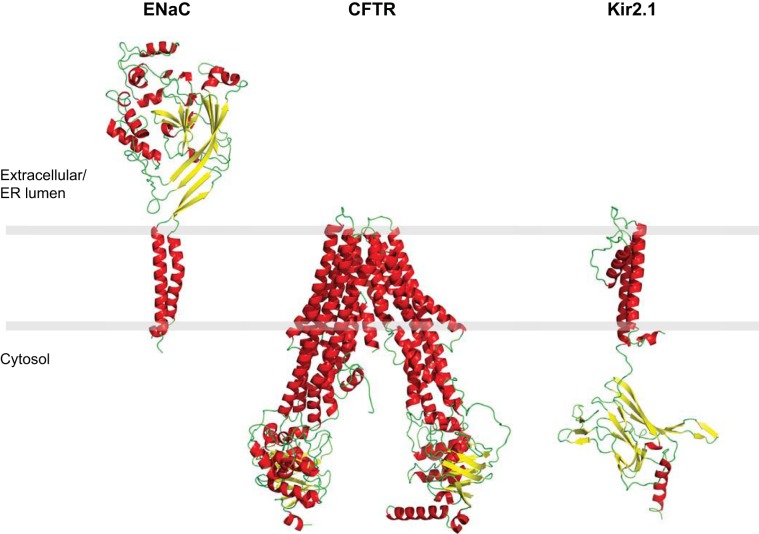
Fig. 1.
Structural models of αENaC, Kir2.1, and CFTR. To generate the structural models, the amino acid sequence of each protein was modeled onto the structures of homologs using the Phyre2 server (76). Protein domains absent from the template structure are not modeled. Specifically, 1,185 amino acids of CFTR (80% of sequence) were modeled onto the multidrug resistance protein, pgp-1 (PDB:4F4C) with 100% confidence. Absent regions include amino acids 1–26, 687–845, 896–911, and 1173–1193. The model for αENaC contains 409 amino acids (60% coverage) modeled onto the ASIC1 crystal structure (PDB: 2QTS) with 100% confidence. Absent regions encompass amino acids 1–106, 601–699, and 199–272 and includes both cytosolic tails. The Kir2.1 model contains 318 amino acids (74% of the sequence) modeled onto the inward rectifying potassium channel Kir2.2 (PDB:3JYC) with 100% confidence. Regions absent correspond to amino acids 1–44 and 372–427.