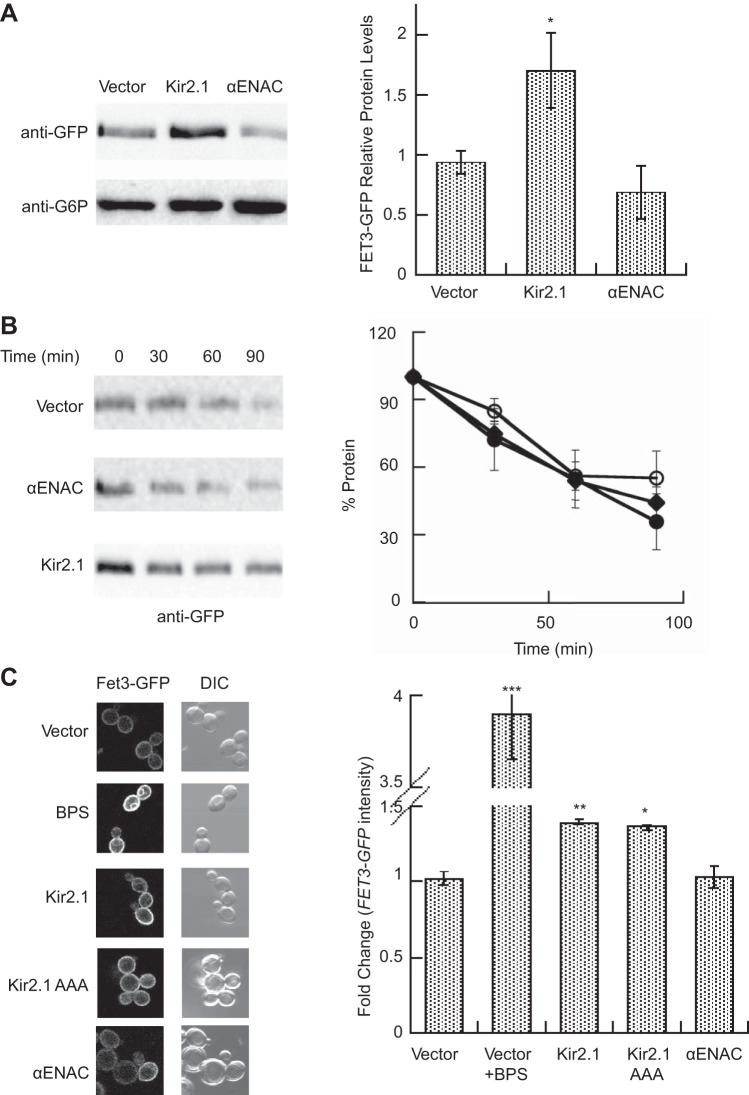
Fig. 6.
Kir2.1 expression increases Fet3 levels and plasma membrane localization. A: lysates of wild-type FET3-GFP yeast transformed with either a vector control or a Kir2.1 or αENaC expression plasmid were immunoblotted with anti-GFP or anti-G6P as a loading control. Data represent the means of 4 experiments ± SE; *P < 0.05 B: cycloheximide chase assays were performed at 30°C as described in materials and methods using the yeast described in A: vector control (closed circles), Kir2.1 expressing (open circles), αENaC expressing (diamonds). Data represent the means of 4–7 experiments ± SE. C: the intensity of plasma membrane fluorescence in the yeast isolates described in A was measured. Data represent the mean values from at least 80 cells, normalized to the average fluorescence of control cells ± SE. Significance was determined by a 1-way ANOVA test with Tukey's post hoc comparison (70); *P < 0.05, **P < 0.01, ***P < 0.0001.