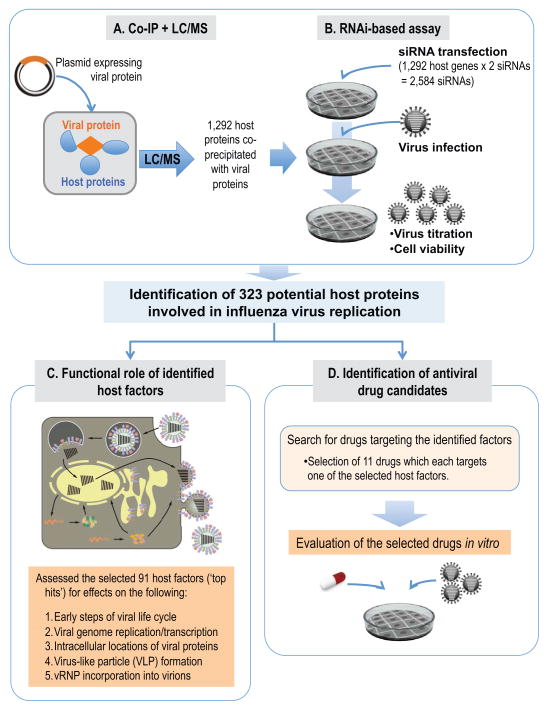
Figure 1. Overview of a systematic study to elucidate the physical and functional host-viral interactions in influenza virus replication, and to identify antiviral drugs.
(A, B) Schematic diagram of the identification of host proteins that co-precipitated with 11 influenza A viral proteins and affected viral replication. (A) Mass spectrometry analysis identified 1,292 host proteins that co-immunoprecipitated with one or more of the 11 FLAG-tagged influenza viral proteins. (B) To identify host factors that affect virus replication, cells were transfected with siRNAs targeted to each of the 1,292 candidate host genes and were then infected with influenza virus. Virus titers and cell viability were then determined. We identified 323 host genes whose mRNA levels were down-regulated, while virus titers were reduced by more than two log10 units compared with a control (299 host factors) or increased by more than one log10 unit (24 host factors). (C) To better understand the role of the identified host factors, we performed mechanistic studies assessing different steps in the viral life cycle for our ‘top hits’, that is, 91 host factors whose siRNA-mediated down-regulation reduced viral replication in cultured cells by at least three log10 units while retaining >80% cell viability. (D) To identify antiviral drugs for influenza virus, we searched for drugs targeting the 299 host factors identified here and selected 11 drugs for in vitro testing. See also Figures S1–4 and Tables S1–3.