Fig. 3.
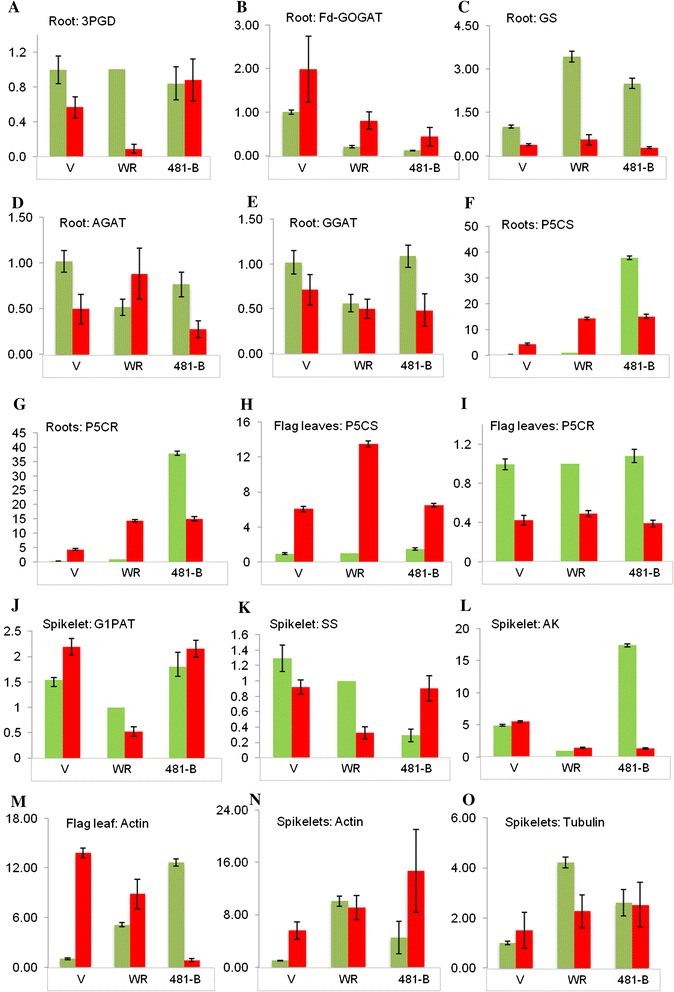
QRT-PCR results for selected amino acid synthesis genes. Genes involved in the metabolic pathways of serine, glutamate, glutamine, glycine, alanine, and proline metabolism were assessed for transcript abundance in the roots of Vandana (V), Way Rarem (WR), and the NIL 481-B. The y axis represents fold change in expression. Bars in green show transcript under well-watered conditions, and those in red indicate abundance under drought. The genes assessed were a 3-phosphoglycerate dehydrogenase (3PGD), b glutamine oxoglutarate aminotransferase (GOGAT), c glutamine synthetase (GS), d alanine glyoxalate aminotransferase (AGAT), e glutamate–glyoxylate aminotransferase (GGAT), f, h pyrroline 5-carboxylate synthetase (P5CS), g, i pyrroline-5-carboxylate reductase (P5CR), j glucose-1-phosphate adenylyltransferase (G1PAT), k sucrose synthase (SS), l adenylate kinase (AK), m, n actin and o tubulin. The average measurement and standard error is shown for each sample (n = 3)
