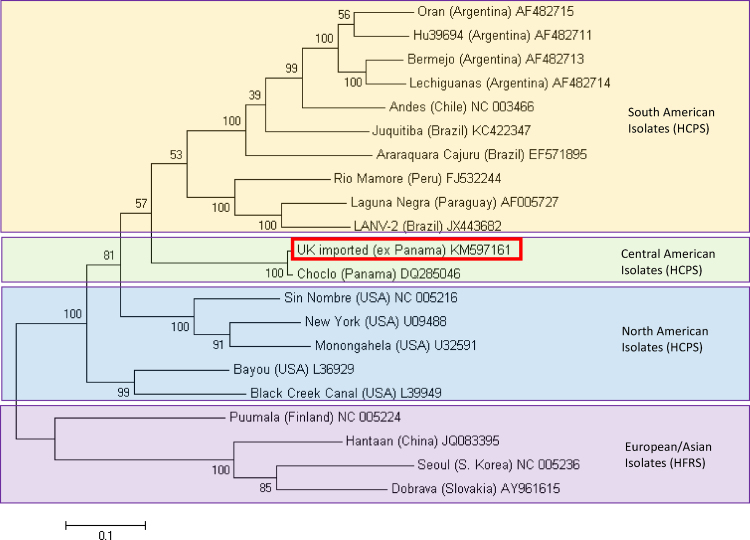
Fig. 2.
Molecular phylogenetic analysis of full S segment hantavirus sequences by maximum likelihood method. The evolutionary history was inferred by using the maximum likelihood method based on the Tamura 3-parameter model conducted in MEGA6. The tree is drawn to scale, with branch lengths measured in the number of substitutions per site. Analysis includes representative isolates for pathogenic ‘New World’ hantaviruses associated with hantavirus cardiopulmonary syndrome (HCPS) and ‘Old World’ hantaviruses associated with haemorrhagic fever with renal syndrome (HFRS). Branch labels include GenBank accession numbers; the sequence highlighted with a red box designates the sequence in this report.