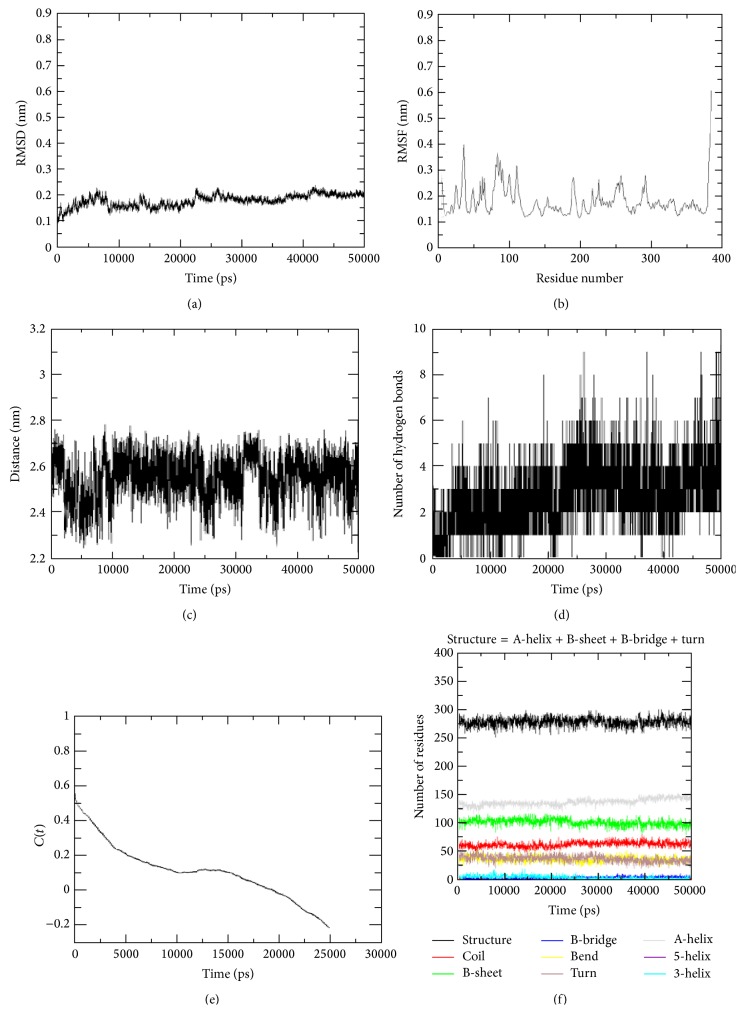
Figure 3.
Stability studies of NBD-p53 motif complex structure. (a) Root mean square deviations (RMSD); (b) backbone atomic fluctuations (RMSF); (c) salt bridge; (d) number of hydrogen bonds; (e) hydrogen bond autocorrelation; and (f) secondary structure analysis of the NBD-p53 motif complex during the 50 ns molecular dynamics simulation.