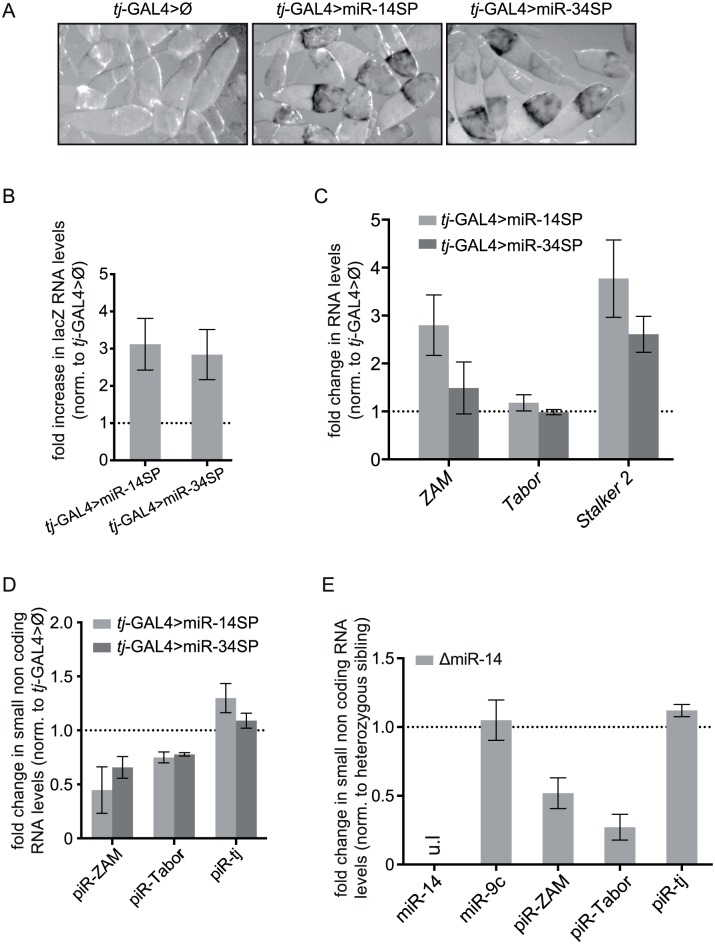
Fig 6. miRNA screen and genetic validation of the requirement of miR-14 for TE repression.
(A) Detection of the piRNA sensor expression following titration of two positive miRNAs by tj-driven expression of the corresponding miRNA-sponges, miR-14SP (middle) and miR-34SP (right). After 1h of β-Gal staining no staining was observed in the sibling ovaries without any miRNA-sponge (Ø), as illustrated by the control for the miR-14SP experiment (left). At that time, only de-repression of the gypsy-lacZ reporter gene, but not yet of the ZAM-lacZ reporter gene, could be detected. (B) Fold changes in the steady-state RNA levels of the lacZ reporters (see primer sequence in S3 Table) upon miR-14SP- and miR-34SP-induced miRNA titration. Quantification was done relative to RpL32 and normalized to sibling ovaries with no miRNA sponge (error bars represent ± SD; n = three biological replicates). (C) Fold changes in the steady-state RNA levels of three follicle cell-specific TEs (ZAM, Tabor and Stalker2) upon miR-14SP and miR-34SP tj-driven expression. Quantification was done relative to RpL32 and normalized to sibling ovaries with no miRNA-sponge (error bars represent the SD of three biological replicates). The absence of Tabor de-repression might indicate that the Tabor family lacks active elements in the tested genotypes. (D) Fold changes in the steady-state level of the three major ZAM, Tabor and traffic jam (tj) piRNAs (see sequences in S3 Table), upon miR-14SP- and miR-34SP-induced miRNA titration. Quantification was done relative to miR-9c and normalized to sibling ovaries with no miRNA sponge (error bars represent the S.D. of three biological replicates). (E) Fold changes in miRNA and piRNA levels induced by the miR-14 null mutation (ΔmiR-14). Quantification was done relative to miR-989 and normalized to heterozygous sibling ovaries. ul: undetectable level (error bars represent the SD of three biological replicates).