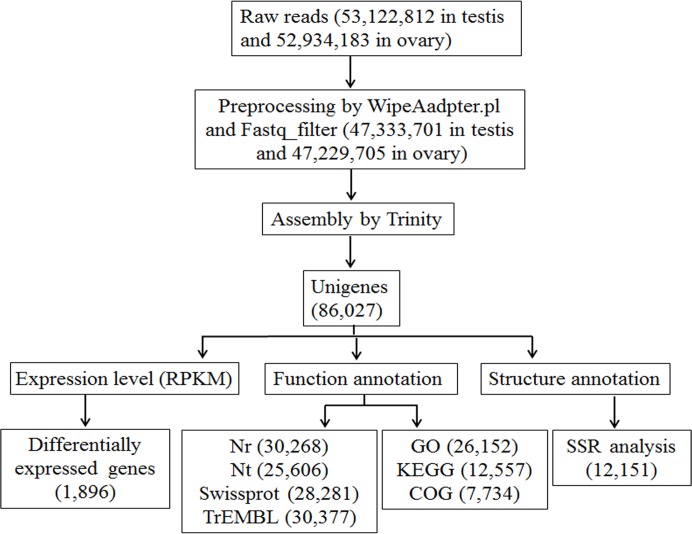
Fig 1. Transcriptome assembly and analysis pipeline.
Clean reads were obtained by pre-processing the raw reads with WipeAadpter.pl software and Fastq_filter software. A total of 86,027 unigenes were acquired by Trinity assembly. The expression levels of the unigenes were normalized with RPKM, through which 1,896 genes differentially expressed between ovary and testis were identified. The unigenes were used for functional annotation and SSR detection.