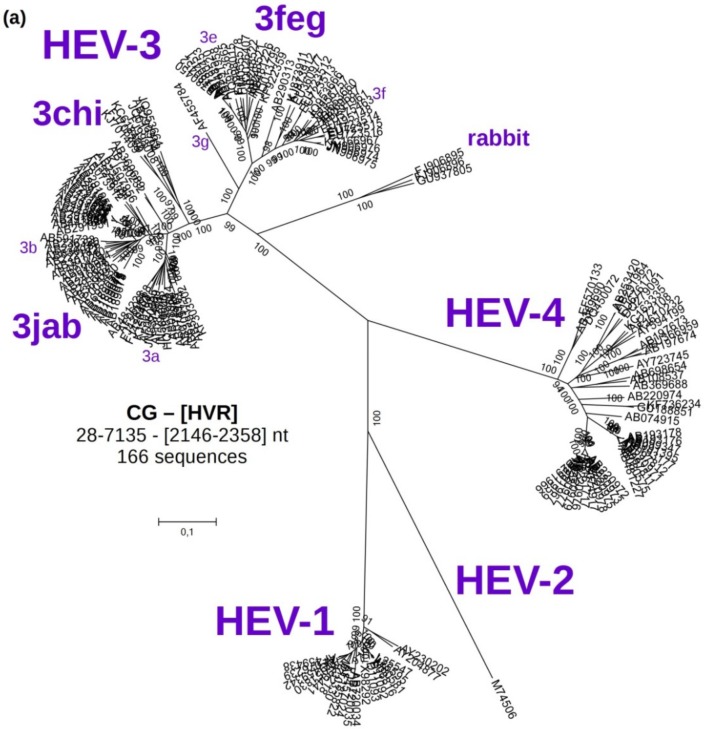
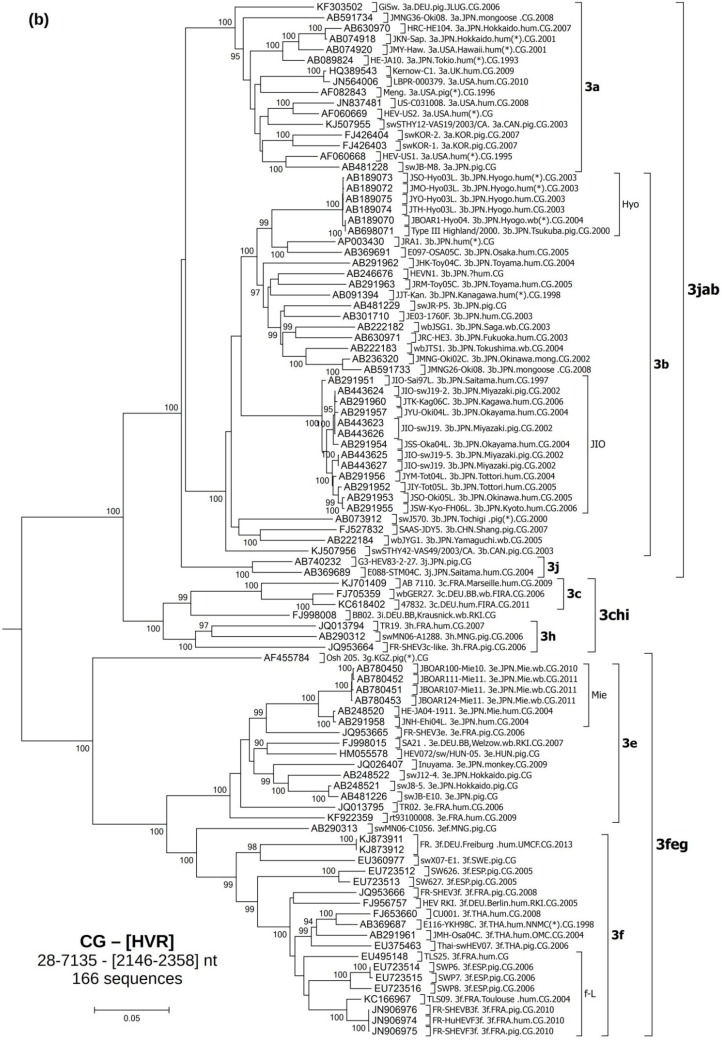
Figure 1.
Molecular Phylogenetic analysis of 166 complete HEV genomes by Maximum Likelihood method based on the Kimura 2-parameter model. The percentage of trees (from 500 bootstrap replicates) in which the associated taxa clustered together is shown next to the branches when over 70%. Initial tree(s) for the heuristic search were obtained by applying the Neighbor-Joining method to a matrix of pairwise distances estimated using the Maximum Composite Likelihood (MCL) approach. A discrete Gamma distribution was used to model evolutionary rate differences among sites (five categories (+G, parameter = 0.5255)). The tree is drawn to scale, with branch lengths measured in the number of substitutions per site. All positions with less than 95% site coverage were eliminated. There were a total of 6868 positions in the final dataset. Evolutionary analyses were conducted in MEGA6. (a) Global view of the unrooted tree, which corresponds to Figure 4 in Lu et al. [32]; (b) Detailed view of the HEV-3 clade. (*)—references sequences cited by Lu; HRC-HE104—strain used in the HEV RNA WHO standard; wb—wild boar. See Supplementary Table 1 for more information.