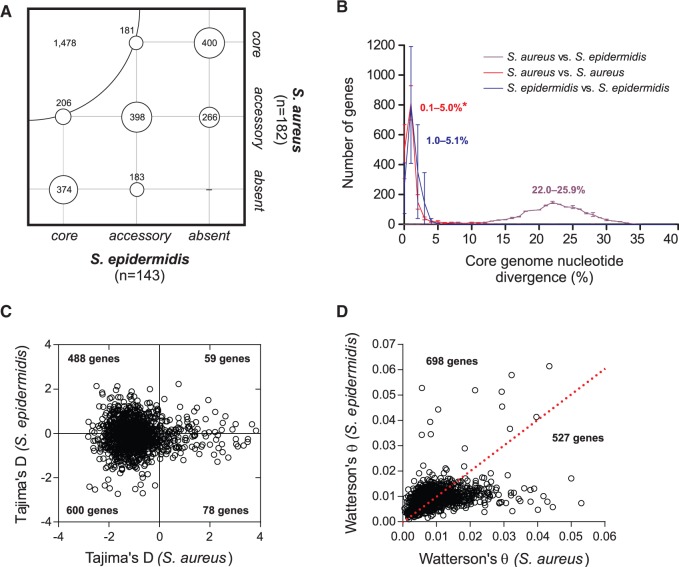
Fig. 2.—
Core and accessory genome variation in S. aureus and S. epidermidis. (A) Overlap between the core and accessory genomes calculated in 324 S. aureus and S. epidermidis genomes. Core genes were defined as being present in 100% isolates, accessory genes in less than 100% but more than 0%, and absent in 0%. The radius of each circle is proportional to the number of detected genes. (B) Core genome nucleotide divergence of representative pairs of S. aureus and S. epidermidis strains between each other. The numbers indicate the range of calculated nucleotide divergence for at least three pairs of strains, and the error bars indicate standard error of the mean. (C) Tajima’s D values for 1,225 core genes shared by both species. The number of genes with different combinations of positive and negative D values is indicated in each quadrant. (D) Watterson’s estimator values for each 1,225 core gene shared by both species. More genes showed a higher θ value in S. epidermidis than in S. aureus.