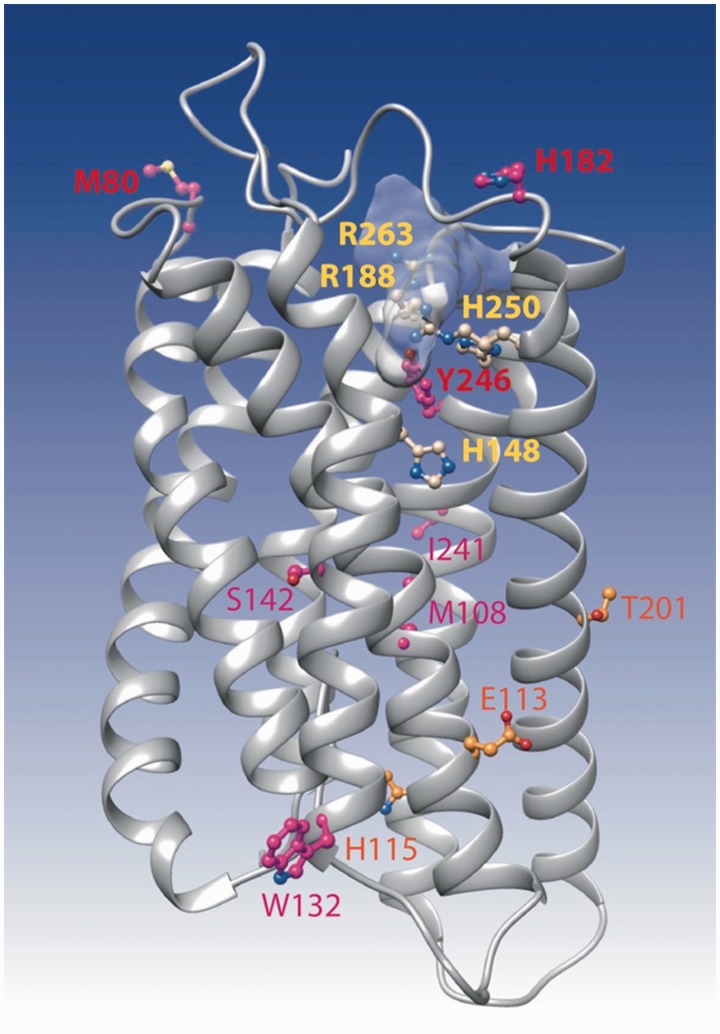
Fig. 8.—
Ribbon representation of the 3D model of FFAR2. The model was made by homology with the 3D structure of the proteinase-activated receptor 1 (pdb 3vw7). This last structure was obtained in complex with the antagonist vorapaxar, whose binding site is shown here light gray shaded. The four critical amino acids (corresponding to H140, R180, H242, and R255 in fig. 3 after correction of the alignment) are presented in yellow, whereas amino acids under positive selection, as reported in table 2, are indicated in orange, red, and purple.