Fig. 1.
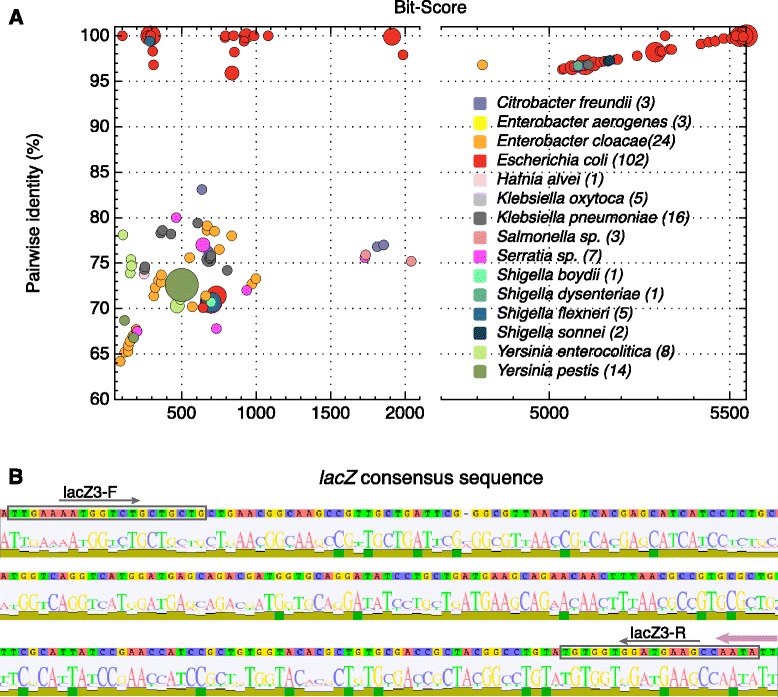
Alignment of lacZ sequences and designing of lacZ3 oligonucleotide primers. a Clusters of DNA sequences hits selected after Blast using lacZ from E. coli K-12 strain MG1655 as the query sequence. 195 hits, with a minimum pairwise identity of 64 % with the query sequence, from different enterobacteria were aligned. The total number of sequences corresponding to each lineage is shown on the right in brackets. The area of each dot correlates with the number of hits from each lineage with the same identity and bit-score. b The consensus sequence of a lacZ fragment (derived from sequences outlined in panel a) was used to design LacZ3-F and LacZ3-R oligonucleotide primers. Consensus and primer sequences were obtained using ClustalW and Primer 3 software, respectively. The degree of conservation of each position in the logo sequence is shown by the relative height of each base. The lacZ3R primer binding site overlaps with 3’ end of the lacZB (Bej et al., [25, 26]) primer (indicated by a pink arrow)
