Abstract
Background:
We showed previously that breast carcinoma amplified sequence 2 (BCAS2) functions as a negative regulator of p53. We also found that BCAS2 is a potential AR-associated protein. AR is essential for the growth and survival of prostate carcinoma. Therefore we characterised the correlation between BCAS2 and AR.
Methods:
Protein interactions were examined by GST pull-down assay and co-immunoprecipitation. Clinical prostate cancer (PCa) specimens were evaluated by immunohistochemical assay. AR transcriptional activity and LNCaP cell growth were assessed by luciferase assay and MTT assay, respectively.
Results:
BCAS2 expression was significantly increased in PCa. BCAS2 stabilised AR protein through both hormone-dependent and -independent manners. There are at least two mechanisms for BCAS2-mediated AR protein upregulation: One is p53-dependent. The p53 is suppressed by BCAS2 that results in increasing AR mRNA and protein expression. The other is via p53-independent inhibition of proteasome degradation. As BCAS2 can form a complex with AR and HSP90, it may function with HSP90 to stabilise AR protein from being degraded by proteasome.
Conclusions:
In this study, we show that BCAS2 is a novel AR-interacting protein and characterise the correlation between BCAS2 and PCa. Thus we propose that BCAS2 could be a diagnostic marker and therapeutic target for PCa.
Keywords: BCAS2, androgen receptor, p53, prostate cancer, HSP90 inhibitor
Breast carcinoma amplified sequence 2 (BCAS2) is a transcriptional cofactor of oestrogen receptor (ER) that enhances oestrogen receptor-α, β (ER-α, β)-mediated gene transcription (Qi et al, 2005) and may involve in breast cancer malignant progression (Worsham et al, 2006). The increased BCAS2 expression coupled with the decreased ERRβ expression was reported in aggressive breast cancer cells (Sengupta et al, 2014). We showed previously that BCAS2 interacts directly with the tumour-suppressor p53 and functions as a negative regulator of p53 in p53-containing cells (Kuo et al, 2009). BCAS2 belongs to the sub-spliceosomal complex containing hPrp19, CDC5L, and PLRG1 (Grote et al, 2010), which has function in pre-mRNA splicing of eukaryotic cell (Chan et al, 2003; Chan and Cheng, 2005; Chanarat and Sträßer, 2013), and contributes in mitotic spindle assembly (Hofmann et al, 2013). Previously, we characterise that BCAS2 is essential for cell growth in Drosophila (Chen et al, 2013). Moreover, the hPrp 19 complex has been shown to participate in the DNA damage response. For example, hPrp19 can interact with terminal deoxynucleotidyl transferase for DNA repair (Mahajan and Mitchell, 2003) and also recruit the DNA repair protein metnase to DNA damage sites (Beck et al, 2008). In addition, CDC5L can directly interact with ATR for the S-phase cell-cycle checkpoint (Zhang et al, 2009). Currently, BCAS2 reportedly binds RPA (a single-strand DNA-binding protein) for the DNA repair function of hPrp19 complex (Wan and Huang, 2014). Thus, BCAS2 seems to be a protein with multiple functions. As we found previously using the yeast two-hybrid system that BCAS2 is a potential androgen receptor (AR)-associated protein (Tsai et al, 2005), we sought to further characterise the correlation between BCAS2 and AR.
AR signalling is essential for prostate carcinoma growth and survival (Taplin, 2008; Saraon et al, 2011; Green et al, 2012; Shafi et al, 2013), and AR protein is existent and function in primary, metastatic, and hormone refractory prostate cancer (Taplin, 2007). Many studies have shown that the progression to castration-resistant prostate cancer is androgen-independent that associates with AR mutation, aberrant AR expression, posttranslational modification, and the altered expression of AR-associated coregulatory proteins (Saraon et al, 2011; Green et al, 2012). Hence, AR is a key molecule in the progression of hormone-dependent and -independent prostate cancer.
AR is a ligand-activated transcription factor (Lavery and Bevan, 2011). In the absence of ligand binding, AR is bound to heat shock protein 90 (HSP90) in the cytoplasm. When androgens bind to AR, they dissociate from HSP90 and translocate to the nucleus to activate the transcription of androgen-regulated genes, including the prostate-specific antigen (PSA) gene, a biomarker for monitoring prostate cancer (Green et al, 2012). Prostate cancer is the most common cancer of males in the developed countries (Jemal et al, 2011). There are several therapeutic regimens for advanced and metastatic prostate cancer, such as androgen deprivation and AR activity elimination (Taplin, 2007; Nacusi and Tindall, 2011). AR activity can be repressed by the tumour-suppressor p53 (Alimirah et al, 2007; Shenk et al, 2001). This study follows our previous demonstration that BCAS2 can suppress p53 protein (Kuo et al, 2009) and is a candidate AR-associated protein (Tsai et al, 2005). Hence, it is of interest to investigate the correlation among BCAS2, p53, and AR in prostate cancer. In this study, we demonstrate that BCAS2 is a novel AR-interacting protein and has strong correlation with AR in clinical prostate cancer samples. Thus, BCAS2, in addition to AR, can be used as a diagnostic marker as well as a therapeutic target in prostate cancer.
Materials and Methods
Plasmids
GST-ARA70 (321–441) was constructed by ligating codons 321–441 of AR-associated protein 70 (ARA70) into the pGEX-4T-1 vector (GE Healthcare, Uppsala, Sweden). The AR and various deletion mutants were constructed by PCR, tagged with HA at their C-termini and are named as indicated (Figure 1F, upper panel). The pcDNA3.1-HSP90-myc plasmid contains HSP90 cDNA in the pcDNA3.1 vector (Invitrogen, Carlsbad, CA, USA). The FLAG-BCAS2, shBCAS2, and shp53 plasmids were described previously (Kuo et al, 2009). pSG5-AR was from Dr Andrew C Cato (Forschungszentrum Karlsruhe, Institute of Genetics, Karlsruhe, Germany) (Shatkina et al, 2003). The PSA reporter construct (PSA-Luc) was kindly provided by Dr Mien-Chie Hung (M. D. Anderson Cancer Center, Houston, TX, USA; Wen et al, 2000).
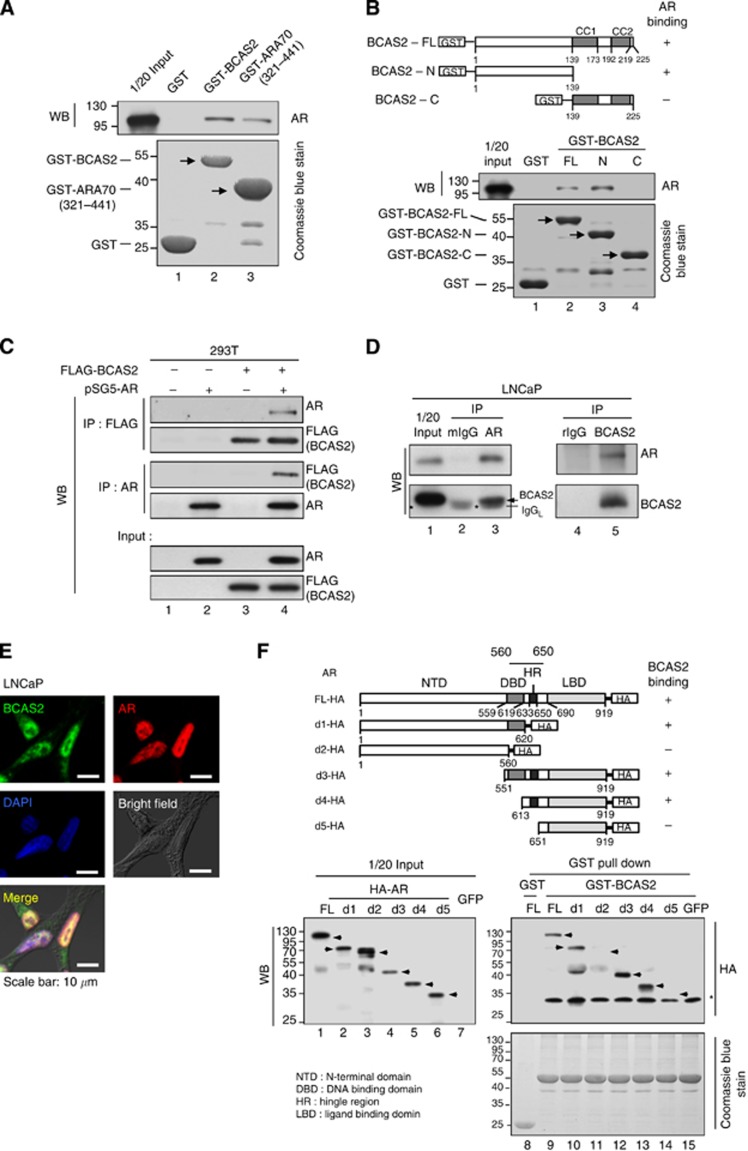
Figure 1.
BCAS2 interacts directly with AR protein in vitro and in vivo. (A) BCAS2 interacts directly with AR protein in vitro. AR protein transcribed and translated in vitro was incubated with GST fusion protein. Upper panel, bound proteins were analysed by anti-AR antibodies. Lower panel, GST proteins. (B) The N-terminal (aa 1–139) domain of BCAS2 binds AR directly. AR protein produced by in vitro transcription/translation was incubated with the indicated GST fusion proteins and then precipitated with GST beads. The precipitated proteins were probed with the anti-AR antibody (middle panel). Lower panel, GST proteins. (C) Reciprocal interaction of AR and BCAS2 in vivo. 293T cells cotransfected with FLAG-BCAS2 and pSG5-AR expression plasmids were harvested for immunoprecipitation with anti-FLAG and anti-AR antibodies, respectively. (D) Endogenous interaction between BCAS2 and AR. Nuclear extracts of LNCaP were subjected to immunoprecipitation with anti-AR anti-BCAS2 antibodies. Bound proteins were immunoblotted with the following antibodies: mIgG: normal mouse IgG; rIgG: normal rabbit IgG; *: mouse IgG light chain, and arrow: BCAS2. (E) Subcellular colocalisation of BCAS2 and AR. Immunofluorescent staining of LNCaP cells, BCAS2 (green), AR (red), and nuclei (blue). Bar, 10 μm. (F) Mapping the domain of AR for BCAS2 binding. Upper panel, schematic representation of the various AR deletion mutants constructed. Middle, cell extracts from various HA-tagged AR mutants were incubated with GST-BCAS2 protein. Bound proteins were subjected to western blotting analysis with anti-HA antibody. Lower panel, GST proteins. Abbreviations: CC1, coiled-coil domain 1; CC2, coiled-coil domain 2.
Cell culture and chemicals
Human HEK 293T (CRL-11268), LNCaP (CRL-1740), and H1299 (CRL-5803) cell lines were obtained from American Type Culture Collection (ATCC, Manassas, VA, USA), and 293FT cell line was purchased from the Invitrogen company. HEK 293T, H1299, and 293FT cells were maintained in Dulbecco's modified Eagle's medium (Life Technologies, Carlsbad, CA, USA) supplemented with 10% fetal bovine serum (FBS; Life Technologies). LNCaP cells were grown in RPMI 1640 (Life Technologies) containing 10% FBS. For dihydroxytesterone (DHT) treatment, cells were maintained in the phenol red-free RPMI 1640 medium (Sigma-Aldrich, St Louis, MO, USA) containing 5% charcoal–dextran-stripped (CDS) FBS (HyClone, Uppsala, Sweden) for 24 h. Chemicals 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide (MTT); 4′,6-diamidino-2-phenylindole (DAPI); 17-allyamino-17-demethoxygeldanamycin (17-AAG); bovine serum albumin (BSA); dithiothreitol (DTT); doxorubicin (Dox); ethylenediaminetetraacetic acid (EDTA); HEPES; magnesium chloride (MgCl2); NP-40; paraformaldehyde; phenylmethanesulfonyl fluoride (PMSF); polybrene; potassium chloride (KCl); potassium hydroxide (KOH); sodium chloride (NaCl); sodium citrate; sodium deoxycholate; and Triton X-100; Tris base and Tween 20 were purchased from Sigma-Aldrich, and dimethyl sulfoxide (DMSO); ethanol (EtOH); glycerol and xylene were purchased from Merck (Darmstadt, Germany). The cycloheximide (CHX) and MG132 were bought from Calbiochem (Darmstadt, Germany); dihydroxytesterone (DHT) from Wako (Tokyo, Japan); and hematoxylin from Surgipath (Buffalo Grove, IL, USA).
Recombinant protein purification and in vitro pull-down assay
The GST and GST fusion proteins were expressed in Escherichia coli BL21(DE3) cells. Further purification was performed using glutathione-Sepharose 4B beads (GE Healthcare) according to the manufacturer's procedures. For in vitro AR protein synthesis, pSG5-AR plasmid was incubated with TNT quick-coupled transcription/translation system (Promega, Madison, WI, USA) master mix. Equal amounts of in vitro synthesised AR protein were incubated with purified GST-BCAS2 and GST-ARA70 (321–441) proteins in IP buffer (50 mM Tris-HCl (pH 8.0), 150 mM NaCl, 2 mM EDTA, 1% Triton X-100). GST was used as a negative control. Interaction proteins were eluted and resolved by SDS–PAGE. Western blotting was performed with rabbit anti-AR (Santa Cruz, Dallas, TX, USA; sc-816) and rabbit anti-GST (Santa Cruz; sc-459) antibodies, respectively.
Transient transfection and luciferase assays
Transient transfections were performed using the ECM 830 Square Wave Electroporation System (BTX Harvard Apparatus, Holliston, MA, USA) for LNCaP cells according to the manufacturers' instruction or using the calcium phosphate method for 293T, 293FT, and H1299 cells. For luciferase assays, cells were transfected with reporter constructs and internal controls, pRL-CMV or plasmids, as indicated. For LNCaP cells, we used the PSA-Luc (5 μg) along with shBCAS2#1 (20 μg) or shCtrl (20 μg) and internal control pRL-CMV (0.1 μg) for the transcriptional activity analysis. For H1299 cells, we used the PSA-Luc (5 μg) along with pSG5-AR (12 μg), FLAG-BCAS2 (8 μg), and internal control pRL-CMV (0.1 μg) for the transcriptional activity analysis. Adding empty vectors equalised the total amount of plasmid per dish. Luciferase activity was detected by the Dual-Glo Luciferase Assay System (Promega). The results of the promoter firefly luciferase activities were normalised by internal control Renilla luciferase activities (pRL-CMV).
Immunoprecipitation and western blotting analysis
For co-immunoprecipitation, cells were cotransfected with plasmids as indicated. Cell lysates harvested in NP-40 lysis buffer (50 mM Tris-HCl (pH 8.0), 150 mM NaCl, 1% NP-40) were subjected to immunoprecipitation with the indicated antibodies. The mixture was incubated with TureBlot anti-mouse Ig (eBioscience, San Diego, CA, USA; 00–8811) or anti-rabbit Ig IP beads (eBioscience; 00-8800). The protein complex was eluted and resolved by SDS–PAGE. For two-step co-immunoprecipitation, H1299 cells were cotransfected with plasmids expressing HA-AR, Myc-HSP90, and FLAG-BCAS2. Cell lysates harvested in IP buffer were incubated with anti-FLAG M2-agarose (Sigma-Aldrich) for 6 h at 4 °C. After centrifugation, the beads were washed with IP buffer, and the FLAG-BCAS2 protein complex was eluted with IP buffer containing 250 mM NaCl and 3xFLAG peptide (300 μg ml−1; Sigma-Aldrich) for 2 h at 4 °C. An untagged BCAS2 was used as a control in the first immunoprecipitation. For the second immunoprecipitation, rabbit anti-HA tag antibody (for AR detection; Abcam, Cambridge, MA, USA; ab9110) or control normal rabbit IgG (rIgG; Santa Cruz; sc-2027) was added to eluates from first immunoprecipitation, followed by the addition of TrueBlot anti-rabbit Ig IP beads. The protein complex was then eluted and resolved by SDS–PAGE. For western blotting, cell lysates were prepared in RIPA buffer (50 mM Tris-HCl (pH8.0), 150 mM NaCl, 1% NP-40, 0.5% sodium deoxycholate). Proteins were separated on SDS–PAGE, transferred to polyvinylidene difluoride membranes (Millipore, Darmstadt, Germany), blotted with specific antibodies, and detected using an ECL Western blotting detection system (GE Healthcare). The following antibodies were used: rabbit anti-AR antibody (Santa Cruz; sc-816), mouse anti-FLAG M2 antibody (Sigma-Aldrich; F3165), rabbit anti-BCAS2 antibody (Proteintech, Chicago, IL, USA; 10414-1-AP), mouse anti-p53 antibody (Santa Cruz; sc-126), mouse anti-β-actin antibody (Sigma-Aldrich; A5441), rabbit anti-poly (ADP-ribose) polymerase (PARP) antibody (Santa Cruz; sc-7150), mouse anti-α-tubulin antibody (Millipore; CP06), rabbit anti-HA tag antibody (Abcam; ab9110), mouse anti-HSP90 antibody (Santa Cruz; sc-69703), mouse anti-Myc antibody (Santa Cruz; sc-40), normal mouse IgG (Santa Cruz; sc-2025), and normal rabbit IgG (Santa Cruz; sc-2027).
Fluorescence microscopy assay
Overnight-cultured LNCaP cells on chamber slides (Nunc, Waltham, MA, USA) were fixed with 4% paraformaldehyde in phosphate-buffered saline (PBS) for 15 min at room temperature (RT). After washing with PBS, the cells were permeabilised with 0.3% Triton X-100 in PBS for 10 min at RT and then blocked with 0.5% BSA in PBS for 30 min at RT. The AR proteins were detected with mouse anti-AR antibody (BD Biosciences, San Jose, CA, USA; 554225), and the BCAS2 proteins were detected with rabbit anti-BCAS2 antibody (Bethyl Laboratories, Montgomery, TX, USA; A300-915A) at RT for 1 h. Cells were then incubated with secondary antibodies, fluorescein isothiocyanate (FITC)-conjugated goat anti-rabbit IgG antibody (Abcam; ab6717) and Alexa Fluor 594-conjugated goat anti-mouse IgG antibody (Invitrogen; A-11005) at RT for 1 h. Nuclei were stained with 0.2 μg ml−1 DAPI in PBS for 5 min at RT. After rinsing with PBS, cells were mounted with Fluoro-Gel (Electron Microscopy Sciences, Hatfield, PA, USA). Fluorescent images were monitored using a Carl Zeiss confocal microscope system (Carl Zeiss, Jena, Germany; LSM 510 META).
Immunohistochemistry (IHC)
Human clinical prostate specimens were obtained from the National Taiwan University Hospital Tissue Bank; and the prostate cancer tissue array (PR953) was purchased from US Biomax, Inc (Rockville, MD, USA). Studies involving these tissues were approved by the Institutional Review Boards at the National Taiwan University, College of Medicine. For IHC staining, the tissue slides were washed in xylene to remove the paraffin and then rehydrated through serial dilutions of alcohol, followed by washings with a solution of PBS (pH 7.2). Treated sections were then placed in a citrate buffer (10 mM Sodium citrate, 0.05% Tween 20, pH 6.0) and boiled for antigen retrieval. The tissue sections were then stained with rabbit anti-BCAS2 antibody (dilution 1 : 500; Proteintech; 10414-1-AP), and the signal was detected using the SuperSensitive Polymer-HRP IHC Detection System (BioGenex, Fremont, CA, USA) according to the manufacturer's instruction. The intensity of protein expression was scored as following: 0=negative or faint brown nuclear staining, 1=intermediate brown nuclear staining, 2=brown nuclear staining, and 3=dark brown nuclear staining. The extent of protein expression was quantified by evaluating the percentage, scored as 0–100%, of the positive stained areas in relation to the entire tumour areas on the slides. The final immunoreactive score was determined by multiplying the intensity score by the extent score, with the minimum score attainable being 0 and the maximum being 300. The cutoff threshold was set as 150 and a score of ⩾150 was considered as high and <150 as low for the statistical analysis.
Real-time quantitative PCR
Total cellular RNAs were extracted using the Trizol reagents (Invitrogen) according to the manufacturer's instructions. Real-time PCR was carried out with a Power SYBR Green PCR Master Mix (Applied Biosystems, Grand Island, NY, USA) according to the manufacturer's protocol. Gene-specific primers used in this study are shown in Supplementary Table S2, and the reactions were analysed in triplicate and normalised to glyceraldehyde-3-phosphate dehydrogenase.
Preparation of cytoplasmic and nuclear extracts
Cells were harvested, lysed in ice-cold buffer A (10 mM HEPES-KOH (pH 7.9), 1.5 mM MgCl2, 10 mM KCl, 2.5 mM DTT, 0.2 mM PMSF, and protease inhibitor), incubated on ice for 10 min, and then vortexed for 10 s. Insoluble nuclei were separated by centrifugation; the supernatant containing the cytoplasmic extracts were removed and placed in a fresh tube. The nuclear fraction was washed with ice-cold buffer A three times and centrifuged at 4 °C. Pellets (nuclear extracts) were then suspended in ice-cold buffer C (20 mM HEPES-KOH (pH 7.9), 1.5 mM MgCl2, 420 mM NaCl, 25% (v/v) glycerol, 0.2 mM EDTA, 0.5 mM DTT, 0.2 mM PMSF and protease inhibitor) and placed on ice for 20 min. The supernatant containing the nuclear extracts were separated by centrifugation.
Generation of recombinant lentivirus
293FT cells were used for generation of lentiviruses. Lentiviruses of pSIN-based vectors were produced by cotransfection with p8.91 and pMD.G into 293FT cells. Viral titre was measured according to the percentage of cells with GFP expression, observed and counted by fluorescence microscopy. Cells were infected at MOI=5–10 with 8 mg ml−1 of polybrene.
Drug sensitivity analysis by MTT assay
Lentivirus-infected cells were seeded at equal amounts in 96-well cell culture plates and incubated overnight. The cells were then treated with various concentrations of 17-AAG. Twenty-four hours later, cell viability was measured by the MTT assay. The MTT assay was performed with the following steps. Cells were incubated with 0.4 mg ml−1 MTT at 37 °C for 4 h. Precipitates were then dissolved in DMSO at 37 °C for 5 min, and the absorbance of the samples at 550 nm was determined by using Power Wave HT microplate spectrophotometer (Biotek, Winooski, VT, USA).
Cell proliferation assay
To analyse the effect of BCAS2 on cell growth, the lentivirus-infected LNCaP cells, as indicated, were seeded at 2 × 104 cells per well in 96-well cell culture plates with phenol red-free RPMI 1640 medium containing 5% CDS. After 24 h hormone starvation, cells were treated with 10 nM DHT or equal volume EtOH (vehicle for DHT) for 24, 48, 72, and 98 h. Cell proliferation was determined by the MTT assay.
Statistical analysis
The statistically significant differences between groups were measured by the Student's t-test or Pearson's chi-square test using GraphPad Prism (version 5.0; GraphPad Software, La Jolla, CA, USA). P-value <0.05 was considered significant.
Results
BCAS2 is a novel AR-interacting protein
BCAS2 is a potential AR-associated protein (Tsai et al, 2005). To further characterise the interaction between BCAS2 and AR in vitro, the AR protein was synthesised in vitro and incubated with GST, GST-BCAS2, and GST-ARA70 (321–441; as a positive control) (Zhou et al, 2002). The results indicate that AR protein bound with BCAS2 (Figure 1A, lane 2) and ARA70 protein (Figure 1A, lane 3). To further verify the binding of BCAS2 to AR, we mapped the AR-binding domains within BCAS2. BCAS2 contains two coiled-coil domains at C-terminal (Kuo et al, 2009); three GST-tagged BCAS2-FL (aa 1–225), BCAS2-N (aa 1–139), and BCAS2-C (aa 139–225) (Figure 1B; upper) were expressed in bacteria, and incubated with the AR protein generated by an in vitro transcription-translation system. A GST pull-down assay located the AR-binding site to the N-terminal region of BCAS2 (Figure 1B; lower). To analyse this interaction in the cellular environment, Figure 1C showed that BCAS2 and AR were bound together in cells (Figure 1C, lane 4). We investigated further the endogenous interaction between BCAS2 and AR in LNCaP cells; the results showed that endogenous AR protein could bind with BCAS2 (Figure 1D, lane 5); and vice versa (Figure 1D, lane 3). Furthermore, immunofluorescence assays showed that endogenous BCAS2 and AR were colocalised predominantly in the nuclei of LNCaP cells, with little in the cytoplasm (Figure 1E). We further determined which domain of AR was responsible for binding BCAS2. The results showed that residues 560–650 of AR, which include the DNA-binding domain and hinge region domain, are responsible for binding BCAS2 (Figure 1F, lanes 10, 12, and 13). In summary, BCAS2 interacts directly with AR, both in vitro and in vivo.
BCAS2 expression is significantly increased in the high Gleason grade of prostate cancers
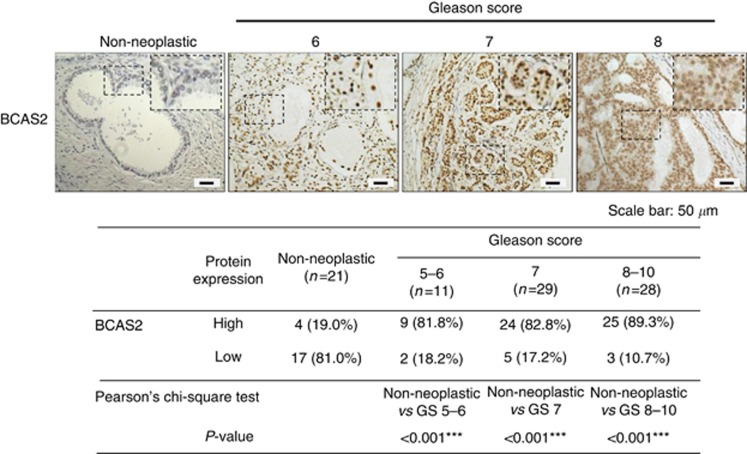
To characterise BCAS2 expression in prostate clinical specimens, 68 prostate cancer specimens and 21 non-neoplastic specimens were assayed by IHC using an anti-BCAS2 antibody. The scores of BCAS2 expression were measured using two parameters (the intensity levels and the percentage of immunoreactive cells) and designated as high or low expression as described in Materials and Methods. The results show that high expression of BCAS2 was only observed in 19% (4/21) of non-neoplastic tissues that we tested. However, high expression of BACS2 was largely detected in the cancerous tissues of 81.8% (9/11) samples with Gleason Score 5–6, 82.8% (24/29) samples with Gleason Score 7, and 89.3% (25/28) samples with Gleason Score 8–10. Representative images (Figure 2) show that BCAS2 was highly expressed in prostate cancer tissues. In sum, these results suggest that BCAS2 expression is upregulated in malignant prostate cancer tissues and BCAS2 expression might be involved in the progression of prostate cancer.
Figure 2.
The BCAS2 expression is significantly increased in high Gleason grade clinical samples. Paraffin-embedded tissue sections of non-neoplastic prostate and prostate cancer specimens were subjected to immunostaining with an anti-BCAS2 antibody. 3,3′-Diaminobenzidine (DAB) was used as a chromogen and hematoxylin as counterstain. Upper panel: left to right represents non-neoplastic prostate, Gleason Score=6, Gleason Score=7, and Gleason Score=8 prostate cancer sections. Lower panel: samples of 68 prostate cancer specimens and 21 non-neoplastic specimens were analysed and scored for the levels of BCAS2 expression. Statistical analysis was done by Peason's chi-square test. Brown, positive staining signals. Blue, hematoxylin counterstain.
BCAS2 enhances AR mRNA and protein expression through the p53-dependent pathway
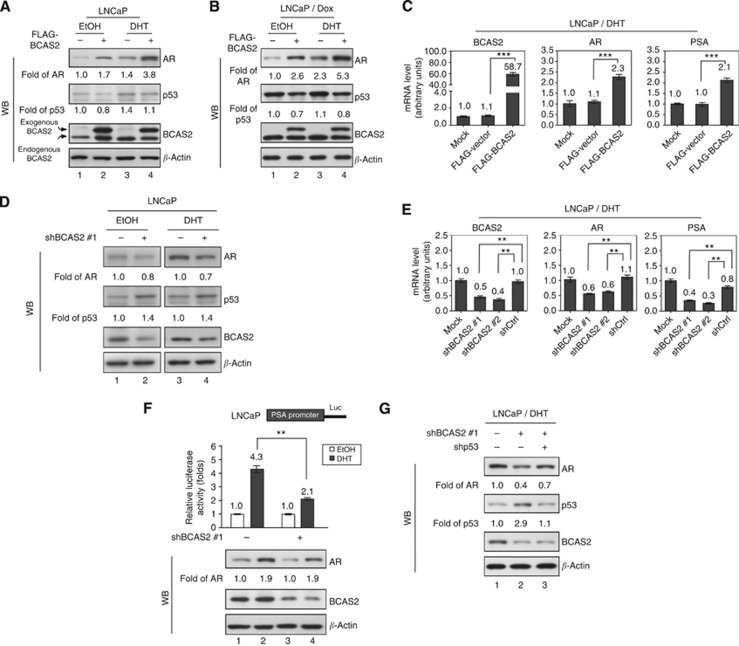
Previous studies have shown that BCAS2 activation can degrade p53 expression (Kuo et al, 2009), and the p53 protein binds to the promoter region of the AR gene, resulting in downregulation of AR expression (Alimirah et al, 2007). These findings imply that AR expression might be increased by BCAS2 through p53 depletion. When enforcing BCAS2 expression in EtOH, the AR increased 1.7-fold (Figure 3A, lane 2) compared with the control (lane 1); DHT increased 1.4-fold (lane 3); and the combination with DHT and BCAS2 could rise to 3.8-fold (lane 4). Meantime, the p53 expression was declined to 0.8-fold in EtOH (lane 2); however, DHT could increase p53 expression to 1.4-fold compared with the control (lane 3). Previously, we found that BCAS2 reduces p53 expression (Kuo et al, 2009), which could be confirmed in lanes 2 and 4 by comparing their corresponding lanes 1 and 3; respectively (Figure 3A). Hence the AR increment in DHT and BCAS2 (lane 4) may partly come from BCAS2-decreasing p53 and in turn increasing AR, the other from DHT that can trigger AR gene expression or more. Interestingly, when comparing p53 expression in either EtOH (lane 1) or DHT treatment (lane 3), DHT could increase p53 gene expression. Consistently, current report demonstrated that low concentration of DHT induces lower mRNA levels in the p53 gene in HNTEP cells; however, high DHT concentration increases the expression of the p53 gene (Pozzobon et al, 2012). To further establish that the increment of AR by BCAS2 is p53 dependent, the BCAS2-transfected LNCaP cells (containing wt p53) were treated with doxorubicin (Dox), a DNA-damaging reagent that is a potent inducer of p53. We then treated LNCaP cells with Dox; the AR and p53 expression profiles (Figure 3B) were similar without Dox treatment (Figure 3A). To analyse lanes 1 and 3 of Figure 3B in which Dox treatment without BCAS2 expression, we could see that p53 mildly increased, but the AR increment could be induced from DHT more than p53. In sum, our data support that DHT can influence AR and p53 gene expression. The reported factors (such as DHT, p53) influence AR expression; now BCAS2 also has a role for AR protein expression. It is very interesting to further know whether DHT can affect BCAS2 gene expression. In this study, we focussed on the relationship between BCAS2 and AR. To extensively examine BCAS2 regulating AR, AR transcription and expression of its downstream gene (PSA) were increased along with the rising BCAS2 mRNA levels (Figure 3C). On the other hand, knockdown of endogenous BCAS2 in cells led to a parallel decrease of AR protein but increase of p53 protein regardless of the presence of ligand (Figure 3D, lanes 2 and 4). The RNAs of AR and PSA decreased along with the depleted BCAS2 RNA caused by shRNAs targeted to BCAS2 (Figure 3E). Two shRNAs against BCAS2 were used: shBCAS2#1, which has been described previously (Kuo et al, 2009), and shBCAS2#2, which was identified in this study (Supplementary Table S1). Analysis of PSA promoter-luciferase activity confirmed that depletion of BCAS2 with shBCAS2#1 repressed PSA promoter activity (Figure 3F, upper panel) coupled with the reduction of AR protein (lower panel), indicating that reducing BCAS2 can diminish AR-transcriptional activity in the presence of DHT. To further confirm that BCAS2 regulation of AR activity is p53-dependent; when both p53 and BCAS2 were knocked down, AR protein levels could be rescued somewhat (Figure 3G, lane 3; and Supplementary Figure S1, lane 3). Taken together, BCAS2 enhances AR mRNA and protein levels through the p53-dependent pathway.
Figure 3.
BCAS2 enhances AR protein expression via a p53-dependent pathway. Overexpression of BCAS2 increases AR levels but decreases p53 protein, regardless of the presence of hormone. Following treatment with DHT (10 nM) or EtOH, LNCaP cells transfected with FLAG-BCAS2 were subjected to western blotting in the absence (A) or presence of 150 nM Dox (B). (C) Overexpression of BCAS2 increases AR and PSA mRNA levels. As shown in panel A, the levels of BCAS2, AR, and PSA mRNA were determined by quantitative PCR. ***: Student's t-test, P<0.001. (D) Reduction of BCAS2 decreases AR and increases p53 protein levels, regardless of the presence of hormone. (E) Depletion of BCAS2 expression reduces AR and PSA mRNA. **: Student's t-test, P<0.01. (F) The deprivation of BCAS2 reduces PSA promoter activity. Upper panel, all luciferase assays were carried out three times in independent experiments. Means±s.d. Lower panel, AR and BCAS2 protein expression. **: Student's t-test, P<0.01. (G) Depletion of BCAS2 decreases AR through a p53-dependent pathway. In the presence of DHT, LNCaP cells transfected with shBCAS2#1 along with shp53 or shCtrl were subjected to WB. The expression levels of AR and p53 protein quantified by UVP BioSpectrum-AC imaging software (UVP, Upland, CA, USA) were normalised to β-actin.
BCAS2 enhances AR protein levels through the p53-independent pathway
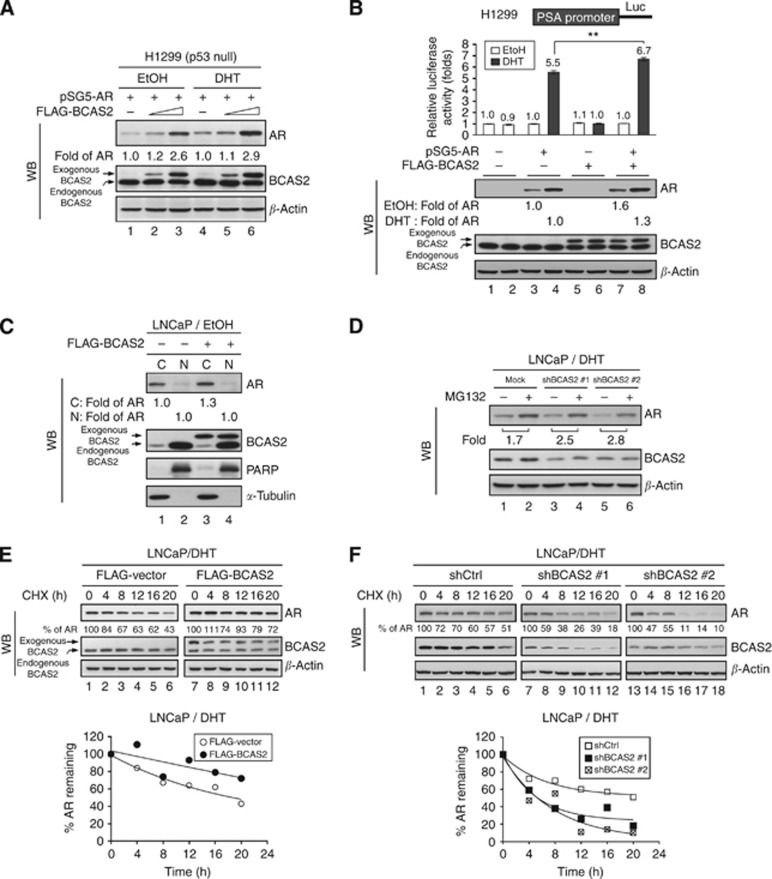
As BCAS2 can interact directly with AR (Figure 1), we speculated that another, p53-independent mechanism may account for the regulation of AR protein levels by BCAS2. Therefore we evaluated whether BCAS2 affected AR protein expression in p53-null cells. H1299 containing AR-null and p53-null cell line and 293T with p53 that is blocked by SV40 T Ag would be used. Hence AR cDNA driven by the heterologous SV40 promoter was transfected into H1299 and 293T along with increasing amounts of the BCAS2 plasmid. The increased AR protein levels were correlated with the amount of BCAS2 expression in H1299 (Figure 4A) and 293T cells (Supplementary Figure S2) regardless of DHT treatment. To analyse further AR transcriptional activity, the luciferase activity of the PSA promoter increased significantly when cotransfected with BCAS2 and AR with ligand (Figure 4B, upper panel, lane 8) and the corresponding AR protein level increased in the presence of BCAS2 with and without ligand treatment (Figure 4B, lower panel, lanes 7 and 8). Although AR protein expression increased in parallel with BCAS2 without ligand treatment, the PSA promoter activity was not affected (Figure 4B, lane 7), indicating that the additional AR protein may be located in the cytosol where it cannot activate gene expression. Therefore we sought to determine whether BCAS2 affects the nuclear translocation of the AR protein and carried out nucleus/cytosol fractionation in the presence of BCAS2 without ligand. The amount of AR protein did not increase in the nucleus in the presence of BCAS2 (Figure 4C lane 4), compared with in the absence of BCAS2 (lane 2), but the amount of AR protein increased in the cytosol (lane 3). Additionally, the endogenous BCAS2 predominately is located in the nucleus; but one-tenth is located in the cytosol from the data of immunofluorescence (Figure 1E) and nucleus/cytosol fractionation (Figure 4C). However, when ectopic transfected BCAS2 plasmid DNA into cells, the amount of BCAS2 equally distributes into nucleus and cytosol (Figure 4C). It may be explained that the overexpressing BCAS2 protein reaches to the saturated condition for shuttling to nucleus. In summary, BCAS2 enhances AR protein levels via a p53-independent mechanism but has no effect on AR nuclear translocation.
Figure 4.
BCAS2 increases AR protein expression via a p53-independent proteasome pathway and enhances AR protein stability. (A) BCAS2 enhances AR protein expression in p53 null cells. H1299 cells were transiently transfected with AR along with increasing doses of FLAG-BCAS2 plasmid. (B) Overexpression of BCAS2 increases AR-dependent transcriptional activity. Upper, luciferase assays. Lower, WB. **: Student's t-test, P<0.01. (C) BCAS2 has no effect on AR nuclear translocation. Without ligand treatment, LNCaP cells overexpressing BCAS2 were harvested for N/C fractionation. Blots were probed for AR, BCAS2, PARP (for the nuclear fraction control), and α-tubulin (cytosolic fraction). Data from nuclear fractions were normalised to PARP; cytosolic fractions were normalised to α-tubulin. (D) BCAS2 protects AR from proteasome degradation. LNCaP cells transfected with shBCAS2#1 or shBCAS2#2 were treated with MG132 (20 μM) or equal volume DMSO (vehicle for MG132) in the presence of DHT. (E) Overexpression of BCAS2 increases AR protein stability. Cells transfected with FLAG-BCAS2 were incubated with CHX (200 μg ml−1) for the times indicated. The percentage of AR indicates the amount of AR at each time point, relative to the control (time 0 set as 100%). Upper panel, WB. Lower panel, the calculated regression curves. (F) Depletion of BCAS2 reduces AR protein stability. LNCaP cells transfected with shCtrl or two different shBCAS2 constructs in the presence of DHT were subjected to CHX-chase assay as described in panel E.
BCAS2 protects AR from proteasomal degradation and increases the half-life of AR in the cytosol and nucleus
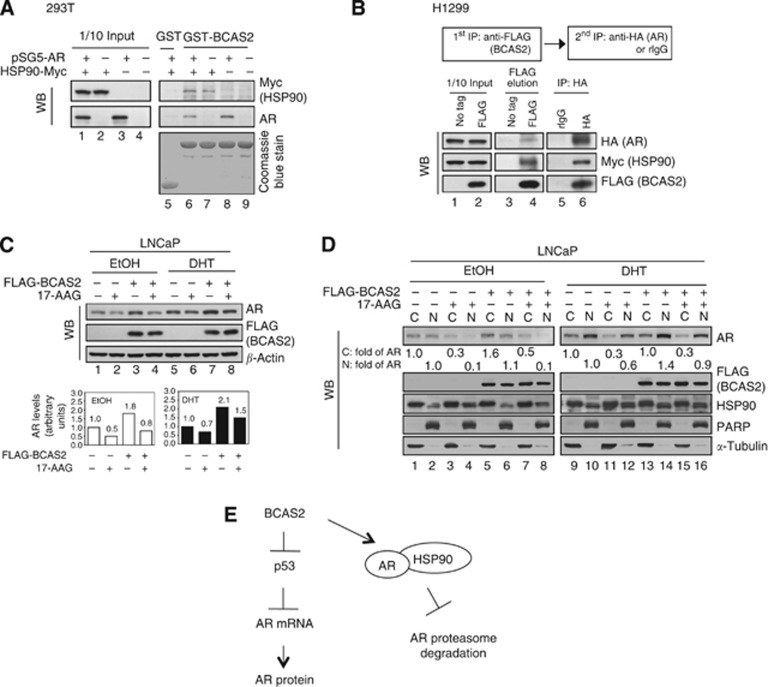
To determine whether BCAS2 is able to protect the AR protein from degradation with treating the proteasome inhibitor, as shown in Figure 4D, in the presence of DHT, MG132 treatment increased the AR protein level 1.7-fold (lane 2) above the control (lane 1). However, AR protein expression increased over 2.5-fold in LNCaP cells treated with two shRNAs of BCAS2 (lanes 3–6). A similar trend was seen in EtOH-treated cells. As shown in Supplementary Figure S3, MG132 treatment produced a 1.8-fold increase in AR protein level (lane 2) compared with the control (lane 1). Nevertheless, the AR protein level increased over 2.3-fold following treatment of the cells with either of the shRNAs (lane 3–6). To examine further whether BCAS2 affects AR protein stability by cycloheximide-chase assay, the half-life of AR in normal condition was about 18 h and BCAS2 could significantly extend the half-life of AR to more than 20 h in the presence of DHT (Figure 4E). In the absence of androgen, the half-life of AR was about 3 h that is consistent with previous report (Lee and Chang, 2003), and BCAS2 prolonged its half-life to 6 h (Supplementary Figure S4A). On the contrary, depletion of endogenous BCAS2 expression in LNCaP cells reduced the half-life of AR protein significantly (about 6 h) compared with the control under treatment with DHT (about 20 h; Figure 4F). But, in the absence of androgen, the half-life of AR was about 1 h when deprived of BCAS2 (Supplementary Figure S4B). Thus BCAS2 expression increases AR protein stability both in the cytosol (no ligand) and in the nucleus (ligand). In summary, BCAS2 has a role in protecting the AR protein from proteasomal degradation and prolongs the half-life of AR protein in the cytosol and nucleus.
BCAS2 can form complexes with HSP90–AR to stabilise the AR protein
HSP90 is a potential BCAS2-binding protein from proteomic assay (Chen et al, 2013) and an essential molecular chaperone for the correct folding and stabilisation of AR (Vanaja et al, 2002; Centenera et al, 2013). We speculated that HSP90 might be involved in BCAS2 stabilising the AR protein. The C-terminal domain of AR has been shown to interact with HSP90 (Marivoet et al, 1992), and our results showed that residues 560–650 of AR, which include the DNA-binding domain and hinge region domain, are responsible for binding BCAS2 (Figure 1F). We then determined whether these three proteins could form complexes. First, in in vitro protein–protein interaction assays, BCAS2 could bind with HSP90 (Figure 5A, lane 7), confirming our previous proteomics assay (Chen et al, 2013), and BCAS2 interacted with AR (Figure 5A, lane 8), consistent with the results in Figure 1. Also, a two-step co-immunoprecipitation assay with H1299 cells co-expressing FLAG-BCAS2, HA-AR, and Myc-HSP90 showed that BCAS2 could bind AR and HSP90 simultaneously (Figure 5B), indicating that BCAS2–AR–HSP90 can form trimetric complex in cells. Furthermore, to determine whether BCAS2 acts in AR protein stabilisation via HSP90, we treated LNCaP cells with 17-AAG in the presence of BCAS2. 17-AAG, an HSP90 inhibitor, can degrade AR protein (Solit et al, 2002). As shown in Figure 5C, overexpression of BCAS2 increased the amount of AR protein under both DHT and EtOH (upper panel, lanes 3 and 7), which is consistent with Figure 3A. But the AR protein level declined 0.5- and 0.7-fold when the cells were treated with 17-AAG along with EtOH and DHT treatment, respectively (upper panel, lanes 2 and 6). However, BCAS2 could rescue the AR protein degradation induced by 17-AAG with DHT and EtOH treatment by 0.8- and 1.5-fold, respectively (upper panel, lanes 4 and 8). To further see the effect of BCAS2–AR–HSP90 complex on AR transcriptional activity, AR-targeted gene (PSA) was measured in the presence of DHT. The increased PSA mRNA was expected owing to BCAS2-stabilising AR (Supplementary Figure S5, lane 3). When treated with 17-AAG, PSA mRNA declined (lane 2); however, BCAS2 could rescue PSA mRNA with 17-AAG treatment (lane 4). Hence, 17-AAG can break the complex and in turn can decrease PSA expression. We further conducted N/C fraction. As shown in Figure 5D, in the absence of ligand, BCAS2 could increase cytosol AR protein 1.6-fold (lane 5) compared with the control, and BCAS2 could mildly rescue 17-AAG-degradating AR protein in the cytosol (compare lane 7 with lane 3). In the presence of DHT, BCAS2 could increase about 40% nuclear AR protein (compare lane 14 with lane 10), and BCAS2 also could rescue 17-AAG-disrupting AR protein in the nucleus (compare lanes 12 and 16). Also HSP90 presented in both the nucleus and cytosol. In sum, BCAS2 can stabilise AR both in the cytosol and nucleus via HSP90-dependent pathway. Taken together, BCAS2 may have a role as a co-chaperone for AR protein stability in HSP90 chaperone complexes.
Figure 5.
BCAS2 can form complexes with HSP90–AR to stabilise AR protein. (A) BCAS2 can form a complex with HSP90–AR. 293T cells transfected with AR and HSP90-Myc plasmids were subjected to GST-BCAS2 precipitation assays. (B) Two-step co-immunoprecipitation identifies a BCAS2–AR–HSP0 complex. The procedure of two-step co-immunoprecipitation is outlined in the box in the upper panel. H1299 cells were co-transfected with HA-AR, Myc-HSP90, and FLAG-BCAS2, and cell lysates were first precipitated with anti-FLAG antibody. Samples were eluted with 3 × FLAG peptide and then immunoprecipitated with anti-HA tag antibody. (C) BCAS2 functions in AR protein stabilisation via HSP90 in the cytosol and nucleus. LNCaP cells transfected with FLAG-BCAS2 were treated with 17-AAG (0.2 μM) in the presence or absence of DHT. Upper panel, AR and BCAS2 protein expression. Lower panel, quantitation of AR protein. (D) BCAS2 rescues both the nuclear and cytosol AR protein degradation induced by 17-AAG. As described in panel C, cells were harvested for N/C fractionation. Fraction samples were subjected to WB analysis for AR, FLAG (BCAS2), HSP90, PARP, and α-tubulin. The expression level of AR protein was quantified and normalised by α-tubulin for the cytosolic fraction and by PARP for the nuclear fraction. (E) Proposed models of BCAS2 increasing AR protein levels.
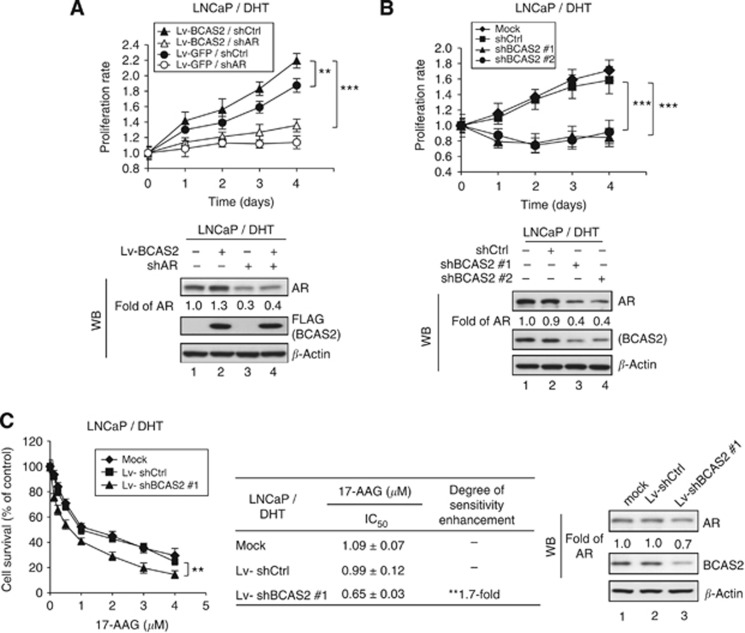
BCAS2 RNAi decreases the cell growth rate via AR degradation and increases sensitivity to a HSP90 inhibitor used to treat prostate cancer
Numerous studies show that knockdown of AR expression decreases the growth rate of LNCaP cells (Cheng et al, 2006; Chang et al, 2012). Because BCAS2 increases the AR protein level, we speculated that BCAS2 might affect the growth of AR-dependent cancer cells. As shown in Figure 6A, overexpression of BCAS2 enhanced the amount of AR protein (lower panel, lane 2) and significantly increased the growth rate of LNCaP cells compared with the control (upper panel). When AR was knocked down, the amount of AR protein was reduced (lower panel, lanes 3 and 4), coupled with a decreased growth rate of LNCaP cells overexpressing BCAS2, and of the GFP control cells (upper panel), indicating that the increased growth rate associated with BCAS2 may be connected to the increased expression of AR. However, the growth rate of LNCaP cells under EtOH conditions was not significantly affected by overexpression of BCAS2 (Supplementary Figure S6A). This can be explained that BCAS2 can increase AR protein levels in the cytosol (Figure 4C, EtOH treatment) but has no effect on the nuclear translocation of AR (Figure 4C), assuming that only nuclear AR can activate the gene expression that is responsible for cell growth. On the other hand, when endogenous BCAS2 expression was reduced in LNCaP cells along with DHT, the growth rate decreased (Figure 6B, upper panel) coupled with a decline in the amount of AR protein (lower panel, lanes 3 and 4). It further supports the linkage of BCAS2 and AR protein levels and their involvement in the growth of LNCaP cells. However, the growth rate of LNCaP cells was also significantly reduced by BCAS2 shRNA treatment without ligand (Supplementary Figure S6B), indicating that, besides AR, BCAS2 also may regulate other molecules, such as p53 and RNA splicing factors (Kuo et al, 2009; Chen et al, 2013).
Figure 6.
BCAS2 affects the growth rate of LNCaP cells via an AR-dependent pathway and the sensitivity of LNCaP cells to a HSP90 inhibitor. (A) Overexpression of BCAS2 increases the growth rate of prostate cancer cells expressing AR. LNCaP cells infected with Lv-BCAS2 or Lv-GFP were then transfected with shAR or shCtrl. Upper panel, the growth rates of LNCaP cells were monitored using the MTT assay. Lower panel, AR and BCAS2 proteins. **: Student's t-test, P<0.01; ***: Student's t-test, P<0.001. (B) Depletion of BCAS2 reduces the growth rate. The growth rates of LNCaP cells transfected with shBCAS2#1, shBCAS#2, or shCtrl plasmids were also determined using the MTT assay. (C) BCAS2 RNAi can increase sensitivity to the HSP90 inhibitor. LNCaP cells infected with Lv-shBCAS2#1 or Lv-shCtrl were treated with various doses of 17-AAG. Cell proliferation was then measured using the MTT assay (left panel), and cell lysates were collected for western blotting analysis (right panel). **: Student's t-test, P<0.01.
HSP90 has been considered as a target for cancer therapy (Solit et al, 2003; Solit and Rosen, 2006; Usmani et al, 2009) and the HSP90 inhibitor 17-AAG is a promising antitumour agent currently undergoing clinical trials for cancer therapy (Porter et al, 2010; Jhaveri et al, 2012). Therefore, we sought to determine whether silencing BCAS2 could increase the 17-AAG sensitivity when cells were expressing the AR protein. As shown in Figure 6C, the cell survival rate was measured by MTT assay in different doses of 17-AAG (left panel). The IC50 value of 17-AAG decreased 1.7-fold in Lv-shBCAS2#1-treated cells compared with the controls (middle panel) with a corresponding decrease of BCAS2 and AR proteins (right panel, lane 3), indicating that depletion of BCAS2 mildly increases sensitivity to 17-AAG in prostate cancer.
Discussion
In this study, we show that BCAS2 is a novel AR-interacting protein (Figure 1), and its expression is significantly increased in high Gleason grade of prostate cancer (Figure 2). AR is a key factor in the progression of hormone-dependent and -independent prostate cancers (Taplin, 2007, Taplin, 2008; Saraon et al, 2011; Green et al, 2012; Shafi et al, 2013). Recently, several AR-binding proteins have been identified as potential diagnostic markers of prostate cancer, for example, ARA55 and Stat5 (Rahman et al, 2004; Thomas et al, 2011). Similarly, the expression of Stat5 is reportedly higher with Gleason score in prostate cancers (Li et al, 2004; Tan et al, 2008; Thomas et al, 2011). The Stat5 is also an AR-interacting protein and functions to stabilise AR protein (Tan et al, 2008; Koptyra et al, 2011). Additionally, AR protein stabilisation/degradation are reportedly correlated with prostate cancer. Several reports demonstrating the mechanisms of AR protein stability/degradation; for example, AR can be degraded via MDM2 E3 ligase (Lin et al, 2002; Gaughan et al, 2005). On the other hand, AR can interact with PTEN, exposing the active site of AR for recognition by caspase-3, leading to AR cleavage and degradation (Lin et al, 2004). In addition, with exposure to an environmental toxin (dioxin), AR can be degraded via DDB1–CUL4B E3 ligase (Ohtake et al, 2011). Our previous report showed that the damaged DNA-binding protein 2 (DDB2) also is an AR-interacting protein. Following contact with AR, the CUL4A–DDB1 complex interacts with DDB2-AR, leading to AR ubiquitination/degradation under stress conditions (Chang et al, 2012). Here we demonstrate that BCAS2 functions in AR protein stabilisation regardless of the presence of hormone. Now, we can add BCAS2 as a new AR-binding protein to stabilise AR, which is involved in the development of prostate cancer and its progression.
BCAS2 is predominately located in the nucleus and one-tenth of BCAS2 is in cytosol (Figure 4C, lanes 1 and 2) as confirmed by immunofluorescence assay (Figure 1E). As to AR, from biochemical N/C fractionation of LNCaP cells, AR presented in both the cytosol and nucleus in the absence of hormone (Figure 5D, lanes 1 and 2), but higher amount of AR presented in the nucleus in the presence of ligand (Figure 5D, lanes 9 and 10). Hence, BCAS2 interaction with AR to increase AR protein can occur both in the cytosol and nucleus (Figure 3). But when the BCAS2–AR complex is in the cytosol, it could not activate PSA promoter activity that can be supported from the data of Figure 4B in which the BCAS2-increased AR could not activate PSA promoter activity without DHT condition (Figure 4B, lane 7). Here we illustrate at least two mechanisms through which BCAS2 increases the amount of AR protein (Figure 5E). The first is via p53-dependent transcription, and our results are in accord with the scenario of p53 reducing AR expression because BCAS2 degrades p53 protein (Kuo et al, 2009), which normally causes an increase of AR mRNA and protein (Alimirah et al, 2007). The second mechanism through which BCAS2 increases AR protein levels is by p53-independent inhibition of proteasome degradation. Since BCAS2–AR–HSP90 can form a tri-complex (Figure 5B), and BCAS2 binds with AR via its DNA binding and hinge region domains (Figure 1F) and HSP90 binds with the C domain of AR (Marivoet et al, 1992). We speculate that BCAS2 and HSP90 may interact with AR collaterally. HSP90 is reportedly required to maintain the correct conformation of AR protein for its stabilisation (Vanaja et al, 2002; Centenera et al, 2013) and BCAS2 seems to function as a co-chaperone in the HSP90 complex to assist AR stabilisation. This is supported by the data showing that overexpression of BCAS2 can rescue AR protein from degradation induced by HSP90 inhibitor under EtOH and DHT treatments (Figure 5C). Furthermore, when N/C fraction was conducted, overexpressing BCAS2 could rescue the degradation of AR caused by HSP90 inhibitor both in the cytosol and nucleus regardless of ligand. In addition, HSP90 presented in both the nucleus and cytosol (Figure 5D). It indicates that BCAS2 can stabilise AR both in the cytosol and nucleus via HSP90-dependent pathway. Recent reports demonstrate that glucocorticoid receptor (GR), non-liganded GR–HSP90 complexes, and ligand binding GR–HSP90 are found in a dynamic equilibrium between the cytosol and nucleus and can shuttle continuously across the nuclear membrane (McNally et al, 2000; Pratt et al, 2006; Biddie and Hager, 2009). The mechanism of AR protein stabilisation by the BCAS2–AR–HSP90 is an interesting subject for future investigation.
There are several drugs targeting AR for prostate cancer therapy, such as the HSP90 inhibitor (Solit et al, 2002; Chen et al, 2008; Heath et al, 2008). BCAS2 can be a target for cancer therapy, because it is a diagnostic marker of prostate cancer (Figure 2). Our results show that BCAS2 RNAi-treated LNCaP cells (p53 wt and AR positive) exhibit decreased growth rates (Figure 6B and Supplementary Figure S6B) and that this involves at least two mechanisms: p53 induction to kill cells (Kuo et al, 2009), and AR depletion to reduce cell growth. Hence, the depletion of BCAS2 in prostate cancer may be a good option for prostate cancer treatment. Additionally, we recently also showed that BCAS2 can form hPrp19/CDC5L complexes for pre-mRNA splicing (Chen et al, 2013), indicating that BCAS2 is also involved in other cell growth regulation pathways (Bonnal et al, 2012). Therefore BCAS2 can be a valuable target for developing therapeutic agents for prostate cancer and for other cancers.
Recently, combination drug strategies using RNAi targeted to specific genes, along with anticancer drugs, are promising for cancer treatment (Paduano et al, 2006; Gray et al, 2007; Holmes et al, 2008; Smith et al, 2009). The HSP90 inhibitor-17-AAG is in the phase II clinical trial as an anticancer drug (Heath et al, 2008; Solit et al, 2008; Modi et al, 2011; Pacey et al, 2012). Here we also found that the depletion of BCAS2 can enhance the IC50 sensitivity of 17-AAG in prostate cancer cells (Figure 6C). Our results support using the combination of BCAS2 RNAi and 17-AAG for prostate cancer treatment. Hence, patients overexpressing BCAS2 can be treated with BCAS2 RNAi along with traditional cancer drugs, and the dose of those drugs may be reduced to decrease the toxic effects.
Acknowledgments
This work was supported by the National Science Council Grants 101-2325-B-002-072, 99-3112-B-002-037, 99-2628-B-002-023-MY3, and 98-2321-B-002-016 and the National Taiwan University Grant 99R311001. We thank Dr Jih-Hwa Guh, Dr Shiou-Hwei Yeh, Dr June-Tai Wu, and Dr Jer-Tsong Hsieh for critical discussion. We also thank Dr Tim J Harrison for editing our English.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on British Journal of Cancer website (http://www.nature.com/bjc)
Supplementary Material
References
- Alimirah F, Panchanathan R, Chen J, Zhang X, Ho SM, Choubey D. Expression of androgen receptor is negatively regulated by p53. Neoplasia. 2007;9:1152–1159. doi: 10.1593/neo.07769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck BD, Park SJ, Lee YJ, Roman Y, Hromas RA, Lee SH. Human Pso4 is a metnase (SETMAR)-binding partner that regulates metnase function in DNA repair. J Biol Chem. 2008;283:9023–9030. doi: 10.1074/jbc.M800150200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biddie SC, Hager GL. Glucocorticoid receptor dynamics and gene regulation. Stress. 2009;12:193–205. doi: 10.1080/10253890802506409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonnal S, Vigevani L, Valcarcel J. The spliceosome as a target of novel antitumour drugs. Nat Rev Drug Discov. 2012;11:847–859. doi: 10.1038/nrd3823. [DOI] [PubMed] [Google Scholar]
- Centenera MM, Fitzpatrick AK, Tilley WD, Butler LM. Hsp90: still a viable target in prostate cancer. Biochim Biophys Acta. 2013;1835:211–218. doi: 10.1016/j.bbcan.2012.12.005. [DOI] [PubMed] [Google Scholar]
- Chan SP, Cheng SC. The Prp19-associated complex is required for specifying interactions of U5 and U6 with pre-mRNA during spliceosome activation. J Biol Chem. 2005;280:31190–31199. doi: 10.1074/jbc.M505060200. [DOI] [PubMed] [Google Scholar]
- Chan SP, Kao DI, Tsai WY, Cheng SC. The Prp19p-associated complex in spliceosome activation. Science. 2003;302:279–282. doi: 10.1126/science.1086602. [DOI] [PubMed] [Google Scholar]
- Chanarat S, Sträßer K. Splicing and beyond: the many faces of the Prp19 complex. Biochim Biophys Acta. 2013;1833:2126–2134. doi: 10.1016/j.bbamcr.2013.05.023. [DOI] [PubMed] [Google Scholar]
- Chang SW, Su CH, Chen HH, Huang CW, Tsao LP, Tsao YP, Chen SL. DDB2 is a novel AR interacting protein and mediates AR ubiquitination/degradation. Int J Biochem Cell Biol. 2012;44:1952–1961. doi: 10.1016/j.biocel.2012.07.023. [DOI] [PubMed] [Google Scholar]
- Chen PH, Lee CI, Weng YT, Tarn WY, Tsao YP, Kuo PC, Hsu PH, Huang CW, Huang CS, Lee HH, Wu JT, Chen SL. BCAS2 is essential for Drosophila viability and functions in pre-mRNA splicing. RNA. 2013;19:208–218. doi: 10.1261/rna.034835.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Sawyers CL, Scher HI. Targeting the androgen receptor pathway in prostate cancer. Curr Opin Pharmacol. 2008;8:440–448. doi: 10.1016/j.coph.2008.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng H, Snoek R, Ghaidi F, Cox ME, Rennie PS. Short hairpin RNA knockdown of the androgen receptor attenuates ligand-independent activation and delays tumor progression. Cancer Res. 2006;66:10613–10620. doi: 10.1158/0008-5472.CAN-06-0028. [DOI] [PubMed] [Google Scholar]
- Gaughan L, Logan IR, Neal DE, Robson CN. Regulation of androgen receptor and histone deacetylase 1 by Mdm2-mediated ubiquitylation. Nucleic Acids Res. 2005;33:13–26. doi: 10.1093/nar/gki141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray PJ, Jr, Stevenson MA, Calderwood SK. Targeting Cdc37 inhibits multiple signaling pathways and induces growth arrest in prostate cancer cells. Cancer Res. 2007;67:11942–11950. doi: 10.1158/0008-5472.CAN-07-3162. [DOI] [PubMed] [Google Scholar]
- Green SM, Mostaghel EA, Nelson PS. Androgen action and metabolism in prostate cancer. Mol Cell Endocrinol. 2012;360:3–13. doi: 10.1016/j.mce.2011.09.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grote M, Wolf E, Will CL, Lemm I, Agafonov DE, Schomburg A, Fischle W, Urlaub H, Luhrmann R. Molecular architecture of the human Prp19/CDC5L complex. Mol Cell Biol. 2010;30:2105–2119. doi: 10.1128/MCB.01505-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heath EI, Hillman DW, Vaishampayan U, Sheng S, Sarkar F, Harper F, Gaskins M, Pitot HC, Tan W, Ivy SP, Pili R, Carducci MA, Erlichman C, Liu G. A phase II trial of 17-allylamino-17-demethoxygeldanamycin in patients with hormone-refractory metastatic prostate cancer. Clin Cancer Res. 2008;14:7940–7946. doi: 10.1158/1078-0432.CCR-08-0221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann JC, Tegha-Dunghu J, Drager S, Will CL, Luhrmann R, Gruss OJ. The Prp19 complex directly functions in mitotic spindle assembly. PLoS One. 2013;8:e74851. doi: 10.1371/journal.pone.0074851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes JL, Sharp SY, Hobbs S, Workman P. Silencing of HSP90 cochaperone AHA1 expression decreases client protein activation and increases cellular sensitivity to the HSP90 inhibitor 17-allylamino-17-demethoxygeldanamycin. Cancer Res. 2008;68:1188–1197. doi: 10.1158/0008-5472.CAN-07-3268. [DOI] [PubMed] [Google Scholar]
- Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61:69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- Jhaveri K, Taldone T, Modi S, Chiosis G. Advances in the clinical development of heat shock protein 90 (Hsp90) inhibitors in cancers. Biochim Biophys Acta. 2012;1823:742–755. doi: 10.1016/j.bbamcr.2011.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koptyra M, Gupta S, Talati P, Nevalainen MT. Signal transducer and activator of transcription 5a/b: biomarker and therapeutic target in prostate and breast cancer. Int J Biochem Cell Biol. 2011;43:1417–1421. doi: 10.1016/j.biocel.2011.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo PC, Tsao YP, Chang HW, Chen PH, Huang CW, Lin ST, Weng YT, Tsai TC, Shieh SY, Chen SL. Breast cancer amplified sequence 2, a novel negative regulator of the p53 tumor suppressor. Cancer Res. 2009;69:8877–8885. doi: 10.1158/0008-5472.CAN-09-2023. [DOI] [PubMed] [Google Scholar]
- Lavery DN, Bevan CL. Androgen receptor signalling in prostate cancer: the functional consequences of acetylation. J Biomed Biotechnol. 2011;2011:862125. doi: 10.1155/2011/862125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee DK, Chang C. Endocrine mechanisms of disease: expression and degradation of androgen receptor: mechanism and clinical implication. J Clin Endocrinol Metab. 2003;88:4043–4054. doi: 10.1210/jc.2003-030261. [DOI] [PubMed] [Google Scholar]
- Li H, Ahonen TJ, Alanen K, Xie J, LeBaron MJ, Pretlow TG, Ealley EL, Zhang Y, Nurmi M, Singh B, Martikainen PM, Nevalainen MT. Activation of signal transducer and activator of transcription 5 in human prostate cancer is associated with high histological grade. Cancer Res. 2004;64:4774–4782. doi: 10.1158/0008-5472.CAN-03-3499. [DOI] [PubMed] [Google Scholar]
- Lin HK, Hu YC, Lee DK, Chang C. Regulation of androgen receptor signaling by PTEN (phosphatase and tensin homolog deleted on chromosome 10) tumor suppressor through distinct mechanisms in prostate cancer cells. Mol Endocrinol. 2004;18:2409–2423. doi: 10.1210/me.2004-0117. [DOI] [PubMed] [Google Scholar]
- Lin HK, Wang L, Hu YC, Altuwaijri S, Chang C. Phosphorylation-dependent ubiquitylation and degradation of androgen receptor by Akt require Mdm2 E3 ligase. EMBO J. 2002;21:4037–4048. doi: 10.1093/emboj/cdf406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahajan KN, Mitchell BS. Role of human Pso4 in mammalian DNA repair and association with terminal deoxynucleotidyl transferase. Proc Natl Acad Sci USA. 2003;100:10746–10751. doi: 10.1073/pnas.1631060100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marivoet S, Van Dijck P, Verhoeven G, Heyns W. Interaction of the 90-kDa heat shock protein with native and in vitro translated androgen receptor and receptor fragments. Mol Cell Endocrinol. 1992;88:165–174. doi: 10.1016/0303-7207(92)90021-w. [DOI] [PubMed] [Google Scholar]
- McNally JG, Muller WG, Walker D, Wolford R, Hager GL. The glucocorticoid receptor: rapid exchange with regulatory sites in living cells. Science. 2000;287:1262–1265. doi: 10.1126/science.287.5456.1262. [DOI] [PubMed] [Google Scholar]
- Modi S, Stopeck A, Linden H, Solit D, Chandarlapaty S, Rosen N, D'Andrea G, Dickler M, Moynahan ME, Sugarman S, Ma W, Patil S, Norton L, Hannah AL, Hudis C. HSP90 inhibition is effective in breast cancer: a phase II trial of tanespimycin (17-AAG) plus trastuzumab in patients with HER2-positive metastatic breast cancer progressing on trastuzumab. Clin Cancer Res. 2011;17:5132–5139. doi: 10.1158/1078-0432.CCR-11-0072. [DOI] [PubMed] [Google Scholar]
- Nacusi LP, Tindall DJ. Targeting 5alpha-reductase for prostate cancer prevention and treatment. Nat Rev Urol. 2011;8:378–384. doi: 10.1038/nrurol.2011.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohtake F, Fujii-Kuriyama Y, Kawajiri K, Kato S. Cross-talk of dioxin and estrogen receptor signals through the ubiquitin system. J Steroid Biochem Mol Biol. 2011;127:102–107. doi: 10.1016/j.jsbmb.2011.03.007. [DOI] [PubMed] [Google Scholar]
- Pacey S, Gore M, Chao D, Banerji U, Larkin J, Sarker S, Owen K, Asad Y, Raynaud F, Walton M, Judson I, Workman P, Eisen T. A Phase II trial of 17-allylamino, 17-demethoxygeldanamycin (17-AAG, tanespimycin) in patients with metastatic melanoma. Invest New Drugs. 2012;30:341–349. doi: 10.1007/s10637-010-9493-4. [DOI] [PubMed] [Google Scholar]
- Paduano F, Villa R, Pennati M, Folini M, Binda M, Daidone MG, Zaffaroni N. Silencing of survivin gene by small interfering RNAs produces supra-additive growth suppression in combination with 17-allylamino-17-demethoxygeldanamycin in human prostate cancer cells. Mol Cancer Ther. 2006;5:179–186. doi: 10.1158/1535-7163.MCT-05-0132. [DOI] [PubMed] [Google Scholar]
- Porter JR, Fritz CC, Depew KM. Discovery and development of Hsp90 inhibitors: a promising pathway for cancer therapy. Curr Opin Chem Biol. 2010;14:412–420. doi: 10.1016/j.cbpa.2010.03.019. [DOI] [PubMed] [Google Scholar]
- Pozzobon A, Schneider L, Brum IS. Androgen-modulated p21 and p53 gene expression in human non-transformed epithelial prostatic cells in primary cultures. Int J Mol Med. 2012;30:967–973. doi: 10.3892/ijmm.2012.1082. [DOI] [PubMed] [Google Scholar]
- Pratt WB, Morishima Y, Murphy M, Harrell M. Chaperoning of glucocorticoid receptors. Handb Exp Pharmacol. 2006;172:111–138. doi: 10.1007/3-540-29717-0_5. [DOI] [PubMed] [Google Scholar]
- Qi C, Zhu YT, Chang J, Yeldandi AV, Rao MS, Zhu YJ. Potentiation of estrogen receptor transcriptional activity by breast cancer amplified sequence 2. Biochem Biophys Res Commun. 2005;328:393–398. doi: 10.1016/j.bbrc.2004.12.187. [DOI] [PubMed] [Google Scholar]
- Rahman M, Miyamoto H, Chang C. Androgen receptor coregulators in prostate cancer: mechanisms and clinical implications. Clin Cancer Res. 2004;10:2208–2219. doi: 10.1158/1078-0432.ccr-0746-3. [DOI] [PubMed] [Google Scholar]
- Saraon P, Jarvi K, Diamandis EP. Molecular alterations during progression of prostate cancer to androgen independence. Clin Chem. 2011;57:1366–1375. doi: 10.1373/clinchem.2011.165977. [DOI] [PubMed] [Google Scholar]
- Sengupta D, Bhargava DK, Dixit A, Sahoo BS, Biswas S, Biswas G, Mishra SK. ERRbeta signalling through FST and BCAS2 inhibits cellular proliferation in breast cancer cells. Br J Cancer. 2014;110:2144–2158. doi: 10.1038/bjc.2014.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shafi AA, Yen AE, Weigel NL. Androgen receptors in hormone-dependent and castration-resistant prostate cancer. Pharmacol Ther. 2013;140:223–238. doi: 10.1016/j.pharmthera.2013.07.003. [DOI] [PubMed] [Google Scholar]
- Shatkina L, Mink S, Rogatsch H, Klocker H, Langer G, Nestl A, Cato AC. The cochaperone Bag-1L enhances androgen receptor action via interaction with the NH2-terminal region of the receptor. Mol Cell Biol. 2003;23:7189–7197. doi: 10.1128/MCB.23.20.7189-7197.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shenk JL, Fisher CJ, Chen SY, Zhou XF, Tillman K, Shemshedini L. p53 represses androgen-induced transactivation of prostate-specific antigen by disrupting hAR amino- to carboxyl-terminal interaction. J Biol Chem. 2001;276:38472–38479. doi: 10.1074/jbc.M103652200. [DOI] [PubMed] [Google Scholar]
- Smith JR, Clarke PA, de Billy E, Workman P. Silencing the cochaperone CDC37 destabilizes kinase clients and sensitizes cancer cells to HSP90 inhibitors. Oncogene. 2009;28:157–169. doi: 10.1038/onc.2008.380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solit DB, Osman I, Polsky D, Panageas KS, Daud A, Goydos JS, Teitcher J, Wolchok JD, Germino FJ, Krown SE, Coit D, Rosen N, Chapman PB. Phase II trial of 17-allylamino-17-demethoxygeldanamycin in patients with metastatic melanoma. Clin Cancer Res. 2008;14:8302–8307. doi: 10.1158/1078-0432.CCR-08-1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solit DB, Rosen N. Hsp90: a novel target for cancer therapy. Curr Top Med Chem. 2006;6:1205–1214. doi: 10.2174/156802606777812068. [DOI] [PubMed] [Google Scholar]
- Solit DB, Scher HI, Rosen N. Hsp90 as a therapeutic target in prostate cancer. Semin Oncol. 2003;30:709–716. doi: 10.1016/s0093-7754(03)00346-4. [DOI] [PubMed] [Google Scholar]
- Solit DB, Zheng FF, Drobnjak M, Munster PN, Higgins B, Verbel D, Heller G, Tong W, Cordon-Cardo C, Agus DB, Scher HI, Rosen N. 17-Allylamino-17-demethoxygeldanamycin induces the degradation of androgen receptor and HER-2/neu and inhibits the growth of prostate cancer xenografts. Clin Cancer Res. 2002;8:986–993. [PubMed] [Google Scholar]
- Tan SH, Dagvadorj A, Shen F, Gu L, Liao Z, Abdulghani J, Zhang Y, Gelmann EP, Zellweger T, Culig Z, Visakorpi T, Bubendorf L, Kirken RA, Karras J, Nevalainen MT. Transcription factor Stat5 synergizes with androgen receptor in prostate cancer cells. Cancer Res. 2008;68:236–248. doi: 10.1158/0008-5472.CAN-07-2972. [DOI] [PubMed] [Google Scholar]
- Taplin ME. Drug insight: role of the androgen receptor in the development and progression of prostate cancer. Nat Clin Pract Oncol. 2007;4:236–244. doi: 10.1038/ncponc0765. [DOI] [PubMed] [Google Scholar]
- Taplin ME. Androgen receptor: role and novel therapeutic prospects in prostate cancer. Expert Rev Anticancer Ther. 2008;8:1495–1508. doi: 10.1586/14737140.8.9.1495. [DOI] [PubMed] [Google Scholar]
- Thomas C, Zoubeidi A, Kuruma H, Fazli L, Lamoureux F, Beraldi E, Monia BP, MacLeod AR, Thuroff JW, Gleave ME. Transcription factor Stat5 knockdown enhances androgen receptor degradation and delays castration-resistant prostate cancer progression in vivo. Mol Cancer Ther. 2011;10:347–359. doi: 10.1158/1535-7163.MCT-10-0850. [DOI] [PubMed] [Google Scholar]
- Tsai TC, Lee YL, Hsiao WC, Tsao YP, Chen SL. NRIP, a novel nuclear receptor interaction protein, enhances the transcriptional activity of nuclear receptors. J Biol Chem. 2005;280:20000–20009. doi: 10.1074/jbc.M412169200. [DOI] [PubMed] [Google Scholar]
- Usmani SZ, Bona R, Li Z. 17 AAG for HSP90 inhibition in cancer—from bench to bedside. Curr Mol Med. 2009;9:654–664. doi: 10.2174/156652409788488757. [DOI] [PubMed] [Google Scholar]
- Vanaja DK, Mitchell SH, Toft DO, Young CY. Effect of geldanamycin on androgen receptor function and stability. Cell Stress Chaperones. 2002;7:55–64. doi: 10.1379/1466-1268(2002)007<0055:eogoar>2.0.co;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan L, Huang J. The PSO4 protein complex associates with replication protein A (RPA) and modulates the activation of ataxia telangiectasia-mutated and Rad3-related (ATR) J Biol Chem. 2014;289:6619–6626. doi: 10.1074/jbc.M113.543439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen Y, Hu MC, Makino K, Spohn B, Bartholomeusz G, Yan DH, Hung MC. HER-2/neu promotes androgen-independent survival and growth of prostate cancer cells through the Akt pathway. Cancer Res. 2000;60:6841–6845. [PubMed] [Google Scholar]
- Worsham MJ, Pals G, Schouten JP, Miller F, Tiwari N, van Spaendonk R, Wolman SR. High-resolution mapping of molecular events associated with immortalization, transformation, and progression to breast cancer in the MCF10 model. Breast Cancer Res Treat. 2006;96:177–186. doi: 10.1007/s10549-005-9077-8. [DOI] [PubMed] [Google Scholar]
- Zhang N, Kaur R, Akhter S, Legerski RJ. Cdc5L interacts with ATR and is required for the S-phase cell-cycle checkpoint. EMBO Rep. 2009;10:1029–1035. doi: 10.1038/embor.2009.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou ZX, He B, Hall SH, Wilson EM, French FS. Domain interactions between coregulator ARA(70) and the androgen receptor (AR) Mol Endocrinol. 2002;16:287–300. doi: 10.1210/mend.16.2.0765. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.