FIG 5 .
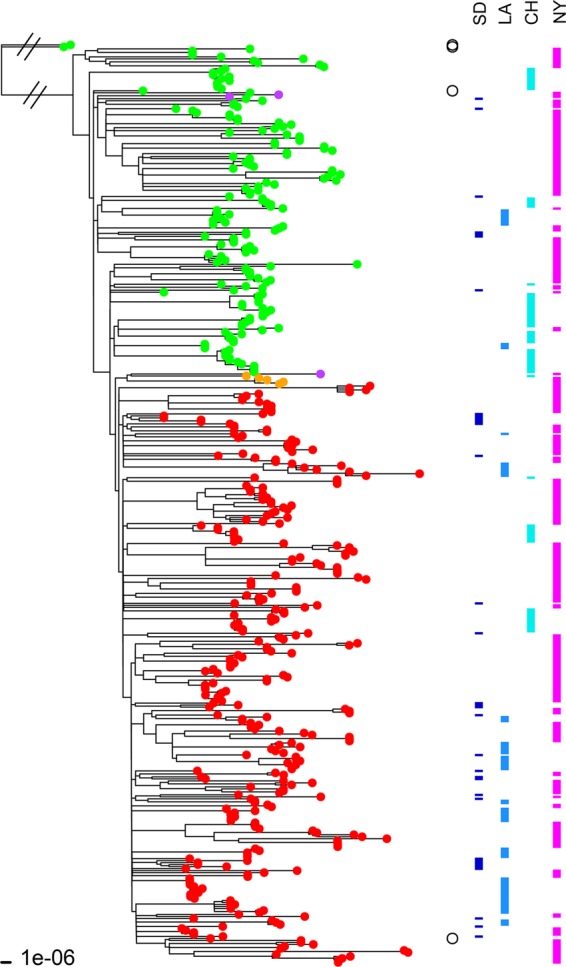
ML tree of 460 USA300 MRSA isolates along with two non-USA300 strains (Newman and COL) included as outgroups. The ML tree was constructed with the 2,104,213-bp core sequence alignment of these isolate genomes in REALPHY. The fluoroquinolone-susceptible isolates (i.e., with no mutations in the grlA and gyrA genes) are indicated by green branch tips, whereas fluoroquinolone-resistant strains (i.e., with the grlA 80Y and gyrA 84L mutations) are indicated by red branch tips. Five fluoroquinolone-resistant isolates had a different set of mutations (grlA 80F and gyrA 84L) and are indicated by orange branch tips. Three isolates indicated by purple tips had only one mutation in the grlA gene (80F). The two non-USA300 strains, Newman (grlA 80S and gyrA 84S) and COL (grlA 80S and gyrA 84S), and two USA300 reference strains, TCH1516 (grlA 80S and gyrA 84S) and FPR3757 (grlA 80Y and gyrA 84L), are indicated by open circles to the right of the tree. USA300 strains TCH1516 and FPR3757 segregated with their fluoroquinolone-susceptible and -resistant clades, respectively. Isolates from San Diego (SD), Los Angeles (LA), Chicago (CH), and New York (NY) are indicated by the bars on the right.
