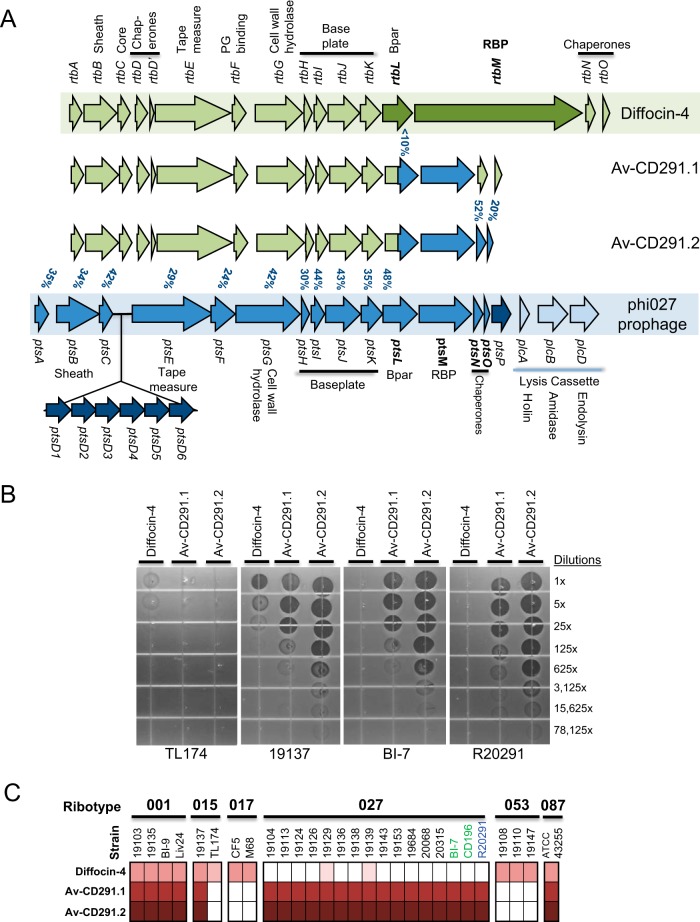
FIG 1 .
Retargeting diffocins with a prophage RBP from C. difficile strain R20291. (A) Schematic representation of gene clusters coding for diffocin-4 (green) and modified diffocins Av-CD291.1 and Av-CD291.2 and including the tail structure genes of the phi027 prophage (blue). Genes are color coded according to source. For the phi027 prophage, the lysis cassette present only in the phi027 prophage is depicted in light blue and structural genes with no homology in the diffocin gene cluster are depicted in dark blue. The percentages of similarity between the diffocin-4 and phi027 genes are given (blue). (B) In vitro spot bioassays for bactericidal activity are shown for several strains. Preparations of diffocin-4, Av-CD291.1, and Av-CD291.2 were serially diluted and spotted on a soft agar lawn containing the indicated target strain. Dark zones of clearance indicate killing. Overlapping but distinct killing specificities for each diffocin preparation, which were all produced from a genetically identical B. subtilis host cell and by the same method, indicate killing is specific to the diffocin and not due to any nonspecific, contaminating B. subtilis protein. (C) The strain coverage for diffocin-4, Av-CD291.1, and Av-CD291.2 for ribotypes 001, 015, 017, 027, 053, and 087 is shown. White indicates no killing, and maroon indicates killing—with intensity of maroon reflecting robustness of killing. Strain designations in green and blue indicate known FQR1 and FQR2 phylogenies, respectively. An additional 20 strains representing 13 ribotypes were also tested and found not to be sensitive to Av-CD291.2 (data not shown).