A tissue-specific cell wall fucogalactoxyloglucan is present in rice.
Abstract
The predominant structure of the hemicellulose xyloglucan (XyG) found in the cell walls of dicots is a fucogalactoXyG with an XXXG core motif, whereas in the Poaceae (grasses and cereals), the structure of XyG is less xylosylated (XXGGn core motif) and lacks fucosyl residues. However, specialized tissues of rice (Oryza sativa) also contain fucogalactoXyG. Orthologous genes of the fucogalactoXyG biosynthetic machinery of Arabidopsis (Arabidopsis thaliana) are present in the rice genome. Expression of these rice genes, including fucosyl-, galactosyl-, and acetyltransferases, in the corresponding Arabidopsis mutants confirmed their activity and substrate specificity, indicating that plants in the Poaceae family have the ability to synthesize fucogalactoXyG in vivo. The data presented here provide support for a functional conservation of XyG structure in higher plants.
The plant cell wall protects and structurally supports plant cells. The wall consists of a variety of polymers, including polysaccharides, the polyphenol lignin, and glycoproteins. One of the major polysaccharides present in the primary walls (i.e. walls of growing cells) in dicots is xyloglucan (XyG), which consists of a β-1,4-glucan backbone with xylosyl substituents. XyG binds noncovalently to cellulose microfibrils and thereby, is thought to act as a spacer molecule, hindering cellulose microfibrils to aggregate (Carpita and Gibeaut, 1993; Pauly et al., 1999a; Bootten et al., 2004; Cosgrove, 2005; Hayashi and Kaida, 2011; Park and Cosgrove, 2012).
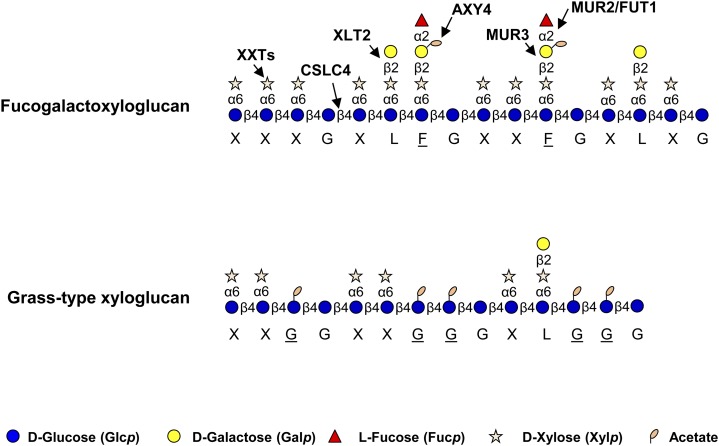
The side-chain substitutions on XyG can be structurally diverse depending on plant species, tissue type, and developmental stage of the tissue (Pauly et al., 2001; Hoffman et al., 2005; Peña et al., 2008; Hsieh and Harris, 2009, 2012; Lampugnani et al., 2013; Schultink et al., 2014). A one-letter code nomenclature has been established to specify the XyG side-chain substitutions (Fry et al., 1993; Tuomivaara et al., 20145). According to this nomenclature, an unsubstituted glucosyl residue is indicated by a G, whereas a glucosyl residue substituted with a xylosyl moiety is shown as an X. In most dicots, the xylosyl residue can be further substituted with a galactosyl residue (L), which in turn, can be further decorated with a fucosyl residue (F) and/or an acetyl group (F/L). In some species, the xylosyl residue can be substituted with an arabinosyl moiety (S), and the backbone glucosyl residue can be O-acetylated (G; Jia et al., 2003; Hoffman et al., 2005).
Numerous genes have been identified in Arabidopsis (Arabidopsis thaliana) that are involved in fucogalactoXyG biosynthesis (Fig. 1; Pauly et al., 2013; Schultink et al., 2014). The glucan backbone is thought to be synthesized by cellulose synthase-like C (CSLC) family proteins, such as AtCSLC4, as shown by in vitro activity data (Cocuron et al., 2007). Several xylosyltransferases (XXTs) from glycosyl transferase family 34 (GT34) are thought to be responsible for XyG xylosylation. Five of these XXTs in Arabidopsis seem to have XXT activity on XyG in vitro (Faik et al., 2002; Zabotina et al., 2008; Vuttipongchaikij et al., 2012; Mansoori et al., 2015). MURUS3 (MUR3) represents a galactosyltransferase that transfers galactosyl moieties specifically to xylosyl residues adjacent to an unsubstituted glucosyl residue on an XXXG unit, converting it to XXLG, whereas Xyloglucan L-side chain galactosyl Transferase2 (XLT2) was identified as another galactosyltransferase transferring a galactosyl moiety specifically to the second xylosyl residue, resulting in XLXG (Madson et al., 2003; Jensen et al., 2012). Both MUR3 and XLT2 belong to GT47 (Li et al., 2004). MUR2/FUCOSYLTRANSFERASE1 (FUT1) from GT37 was found to harbor fucosyltransferase activity, transferring Fuc from GDP-Fuc to a galactosyl residue adjacent to the unsubstituted glucosyl residue (i.e. onto XXLG but not onto XLXG; Perrin et al., 1999; Vanzin et al., 2002). O-acetylation of the galactosyl residue is mediated by Altered Xyloglucan4 (AXY4) and AXY4L, both of which belong to the Trichome Birefringence-Like (TBL) protein family (Bischoff et al., 2010; Gille et al., 2011; Gille and Pauly, 2012).
Figure 1.
Schematic structures of two types of XyGs and known biosynthetic proteins in Arabidopsis (Hsieh and Harris, 2009; Pauly et al., 2013). The corresponding one-letter code for XyG is shown below the pictograms (Fry et al., 1993; Tuomivaara et al., 2015).
XyG found throughout land plants exhibits structural diversity with respect to side-chain substitution patterns (Schultink et al., 2014). Most dicots, such as Arabidopsis, and the noncommelinoid monocots possess a fucogalactoXyG of the XXXG-type XyG structure as shown in Figure 1. However, plant species in the Solanaceae and Poaceae as well as the moss Physcomitrella patens contain a different XyG structure with a reduced level of xylosylation, resulting in an XXGGn core motif (York et al., 1996; Kato et al., 2004; Gibeaut et al., 2005; Jia et al., 2005; Peña et al., 2008; Hsieh and Harris, 2009). In addition, the glucan backbone can be O-acetylated in plants of Solanaceae and Poaceae families (Gibeaut et al., 2005; Jia et al., 2005). XyG from Solanaceae with an XXGG core motif can be further arabinosylated and/or galactosylated (Jia et al., 2005). No XyGs with an XXGGn motif backbone have been reported to be fucosylated.
The function of structural diversity of XyG substitutions, such as fucosylation and/or altered xylosylation pattern, remains enigmatic. Removing the terminal fucosyl or acetyl moieties in the corresponding Arabidopsis mutants does not lead to any change in plant growth and development (Vanzin et al., 2002; Gille et al., 2011). However, removing galactosyl residues as well as fucosyl and acetyl moieties in the Arabidopsis xlt2 mur3.1 double mutant results in a dwarfed plant (Jensen et al., 2012; Kong et al., 2015). Replacing the galactosyl moiety with an arabinofuranosyl residue by, for example, expressing a tomato (Solanum lycopersicum) arabinosyltransferase in the Arabidopsis xlt2 mur3.1 mutant rescues the growth phenotype and restores wall biomechanics, indicating that galactosylation and arabinosylation in XyG have an equivalent function (Schultink et al., 2013). Recently, fucosylated XyG structures were found in the pollen tubes of tobacco (Nicotiana alata) and tomato, indicating that fucogalactoXyG is likely also present in other Solanaceae plants, albeit restricted to specific tissues (Lampugnani et al., 2013; Dardelle et al., 2015). Although there is circumstantial evidence that fucogalactoXyG is present in cell suspension cultures of rice (Oryza sativa) and cell suspension cultures of fescue (Festuca arundinaceae; McDougall and Fry, 1994; Peña et al., 2008), fucogalactoXyG has not been found in any physiologically relevant plant tissues of members of the Poaceae (Kato et al., 1982; Watanabe et al., 1984; Gibeaut et al., 2005; Hsieh and Harris, 2009; Brennan and Harris, 2011). Here, we provide chemical and genetic evidence that fucogalactoXyG is, indeed, present in plant tissues of a grass (rice) and prove that the rice genome harbors the genes that could be part of the synthetic machinery necessary to produce fucogalactoXyG.
RESULTS
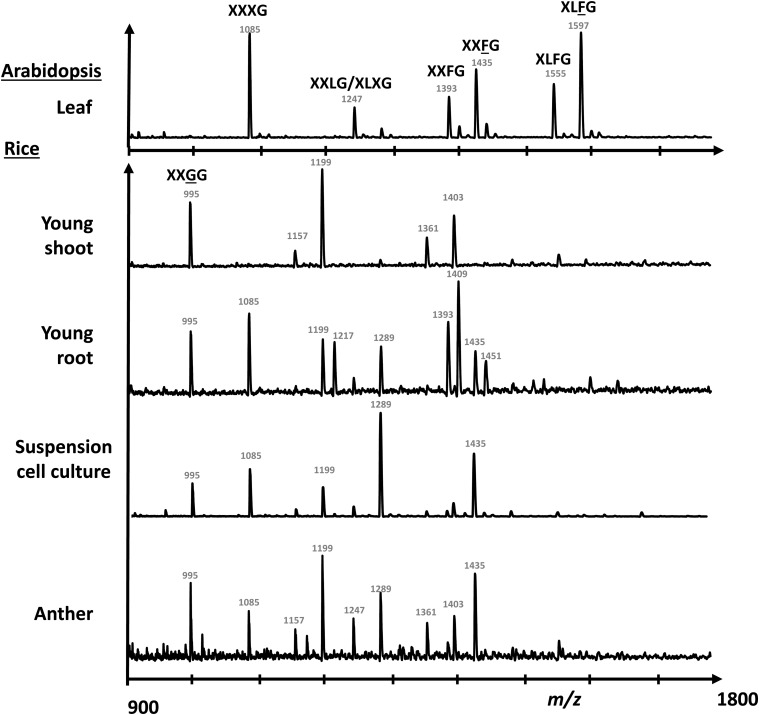
FucogalactoXyG Is Present in Rice Plant Tissues
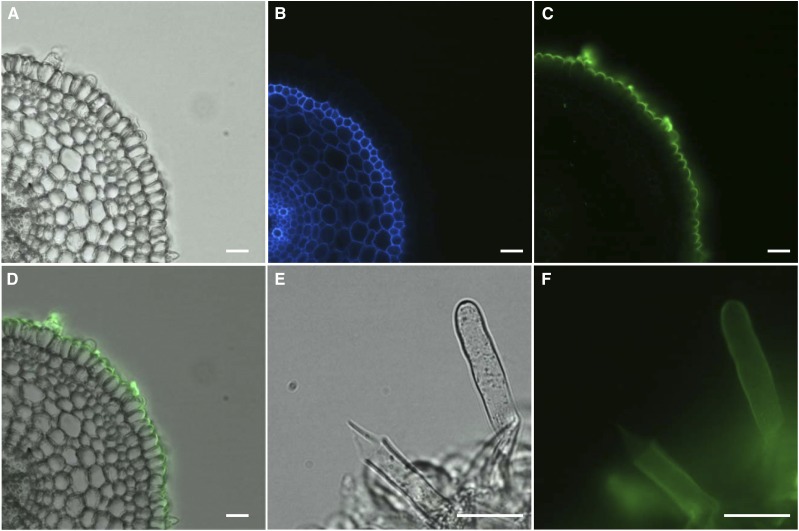
Several types of plant tissues were harvested from rice plants, and their XyG structure was assessed by oligosaccharide mass profiling (OLIMP). XXGGn-type XyG was found to be the predominant XyG in young shoots of rice (Fig. 2), which has been previously reported in other grasses, such as barley (Hordeum vulgare), bamboo (Phyllostachys edulis), and maize (Zea mays; Kato et al., 2004; Gibeaut et al., 2005; Hsieh and Harris, 2009). However, in young root tissue, a variety of XyG structures was detected—not only the XXGGn type but also, oligosaccharide ions, which are consistent with fucogalactosylated XyG oligosaccharides (Fig. 2; Supplemental Table S1). For example, the ion with a mass to charge ratio (m/z) of 1,395 is indicative of an oligosaccharide that consists of five hexoses, three pentoses, and one deoxyhexose. Such an ion is commonly found in XyG oligosaccharide mass profiles in a typical dicot representing XXFG there. These putative fucogalactoXyG structures were also found in rice anthers and wall preparations from a rice suspension cell culture (Fig. 2; Supplemental Table S1). XyG oligosaccharide profiles derived from the rice tissues were also established by high-performance anion-exchange chromatography (HPAEC), showing that a peak with the same retention time as XXFG could be observed (Supplemental Fig. S1). Mass spectrometric analysis of the content of this collected peak confirmed the XyG oligosaccharide m/z of 1,395 (Supplemental Fig. S1). Other oligosaccharides, which are uncommon in dicot XyG oligosaccharide profiles, were also seen in the HPAEC XyG profile. These oligosaccharides likely represent XXGGn-type XyG oligosaccharides, because they represent lowly pentosylated XyG oligosaccharides. XyG glycosyl-linkage analysis from XyG oligosaccharides showed that T-Fucp and 2-Galp, which are typical linkages of fucogalactoXyG, are present in rice young root tissue but notin young shoots (Table I). An increase of 4-Glcp was found in rice young shoots, which is consistent with the OLIMP data that XyG in rice young shoot is less glycosylated. Complex Carbohydrate Research Center Monoclonal Antibody1 (CCRC-M1), an antibody that recognizes a fucosyl-α1,2-galactosyl epitope carried by fucogalactoXyG (Puhlmann et al., 1994; Freshour et al., 2003), was used to confirm the presence of fucosylated XyG in rice and determine its tissue localization. The CCRC-M1 epitope was, indeed, found in rice roots, especially in the outer layer of root epidermal cells as well as root hairs (Fig. 3; Supplemental Fig. S2A). No CCRC-M1 epitope was detected in young shoots of rice (Supplemental Fig. S2D). The epitope of CCRC-M48, galactosylated side chains of nonfucosylated XyG (preferentially XXLG and XLLG), was also found in root but not shoot tissues (Supplemental Fig. S2, B and E), whereas xylan epitopes recognized by Leeds Monoclonal Antibody11 (LM11) were found in vascular tissues in both rice root and shoot (Supplemental Fig. S2, C and F), indicating that fucogalactoXyG seems to be restricted to the outer root epidermis, including root hairs.
Figure 2.
XyG OLIMP of Arabidopsis leaf and various tissues from rice. The m/z of significant ion signals and suggested corresponding structures are labeled. Detailed structural information can be found in Supplemental Table S1.
Table I. Glycosidic linkage analysis of XEG-digested XyG derived from rice shoot and root.
Details can be found in “Materials and Methods.” ND, Not detected; *, statistically significant from shoot (Student’s t test; P < 0.05, n = 3).
| Glycosyl Residue Linkage | Shoot | Root |
|---|---|---|
| T-Fucpa | ND | 1.3 ± 0.2* |
| T-Xylp | 8.0 ± 1.2 | 7.5 ± 0.9 |
| T-Galp | 7.8 ± 3.8 | 7.0 ± 0.3 |
| 2-Xylp | 5.1 ± 2.7 | 6.5 ± 1.2 |
| 2-Galp | ND | 1.9 ± 0.5* |
| 6-Glcp | 24.4 ± 2.0 | 30.1 ± 3.2 |
| 4-Glcp | 39.6 ± 5.2 | 27.1 ± 3.4* |
| 4,6-Glcp | 15.2 ± 3.0 | 18.7 ± 2.8 |
Glycosidic linkages are expressed as percentages of total peak areas.
Figure 3.
Immunofluorescent labeling of 3-d-old rice root transverse sections with CCRC-M1. Root section in bright field (A), autofluorescence (B), CCRC-M1 (C), and merged signal of bright field and CCRC-M1 fluorescent signal (D). Root hairs in bright field (E) and CCRC-M1 fluorescent signal (F). Bars = 25 µm.
Identification of Candidate Genes for the Synthesis of Rice FucogalactoXyG
Because fucogalactoXyG was found in a variety of rice tissues, these tissues should also express the necessary genes to generate this type of XyG. Based on known XyG biosynthetic genes from Arabidopsis, phylogenic trees were constructed for homologs of AtMUR2/FUT1 (XyG fucosyltransferase), AtMUR3, AtXLT2 (XyG galactosyltransferases), and AtAXY4 (XyG acetyltransferase) using rice and Brachypodium genes (Supplemental Figs. S3–S5). To ascertain which of the genes in the trees could represent functional rice XyG transferases, a coexpression analysis was performed with OsCSLC3, the closest homolog of AtCSLC4 thought to be responsible for XyG glucan backbone synthesis in Arabidopsis (Cocuron et al., 2007). Rice orthologs of AtMUR3, AtXLT2, and AtAXY4 were found among the top 50 coexpressed genes (Supplemental Table S2), suggesting that these proteins and homologs in their respective subclade might represent the galactosyl- and acetyltransferases involved in fucogalactoXyG biosynthesis (Table II). There are two coexpressed putative XyG fucosyltransferases among the top 50 coexpressed genes (Supplemental Table S2). However, both proteins were phylogenetically distinct from the MUR2 homologs in Arabidopsis (Supplemental Fig. S3). Therefore, several putative XyG fucosyltransferases were selected for additional characterization (Table I; Supplemental Fig. S3).
Table II. List of candidate genes hypothetically involved in rice fucogalactoXyG biosynthesis.
| Putative Activity and Locus | cDNA | Homologa | Coexpressed with OsCSLC3b |
Cloned | Name Used in This Study | ||
|---|---|---|---|---|---|---|---|
| Array | R Value | Rankc | |||||
| Fucosyltransferase | |||||||
| Os02g52640 | AK101014 | MUR2 | Yes | ||||
| Os02g52630 | AK102499 | MUR2 | Os.49612.1.S1_at | 0.88494 | 34 | Yes | OsMUR2 |
| Os06g10910 | AK073279 | MUR2 | Os.53557.1.S1_at | 0.88705 | 33 | Yes | |
| Os02g52610 | AK072854 | MUR2 | Yes | ||||
| Os02g17534 | AK071664 | MUR2 | Yes | ||||
| Galactosyltransferase | |||||||
| Os04g48480 | AK069178 | XLT2 | Yes | OsXLT2 | |||
| Os06g23420 | AK101619 | XLT2 | Os.54057.1.S1_at | 0.88111 | 36 | ||
| Os03g05060 | AK073950 | MUR3 | Os.53691.1.S1_at | 0.90005 | 20 | ||
| Os03g05110 | AK103041 | MUR3 | Yes | OsMUR3 | |||
| Acetyltransferase | |||||||
| Os07g15230 | None | AXY4 | |||||
| Os09g20850 | None | AXY4 | |||||
| Os06g12870 | AK240937 | AXY4 | Yes | OsAXY4 | |||
| Os07g15320 | None | AXY4 | Os.36470.1.S1_at | 0.88017 | 37 | ||
Homolog of known Arabidopsis protein.
Coexpression data from GeneCAT (Mutwil et al., 2008). Probe-set OsAffx.17009.1.S1_x_at was used as the query.
Rank in coexpression set.
Impact of Rice Candidate Genes on XyG Structures in Arabidopsis
To assess the activity of these candidate genes, a functional complementation approach was used by transforming the candidate genes into the corresponding Arabidopsis mutant lacking the presumed activities. The putative rice fucosyltransferases were expressed in the Arabidopsis axy2.2/fut1/mur2 mutant lacking fucosylated XyG (Supplemental Figs. S6 and S7). The galactosyltransferases were expressed in the Arabidopsis xlt2 mur3.1 double mutant lacking galactosylated XyG, and the acetyltransferase was expressed in the Arabidopsis axy4-3 mutant lacking O-acetylated XyG.
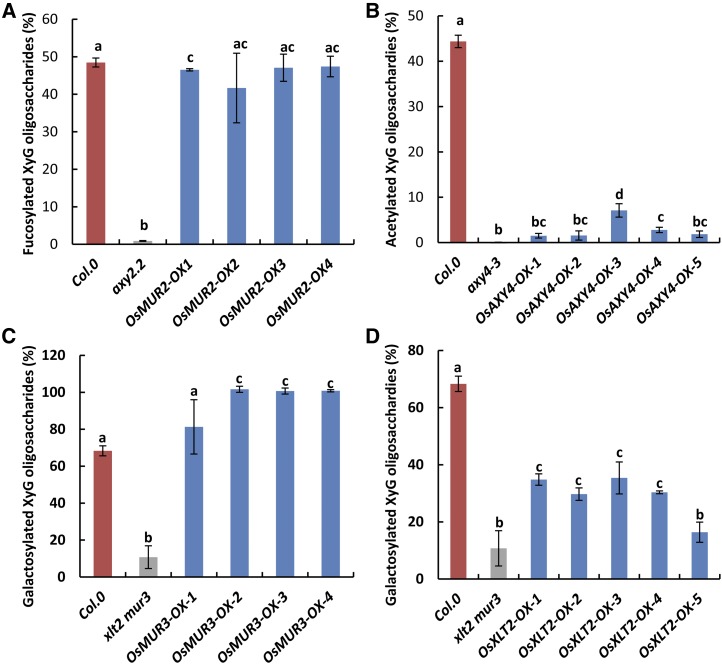
Multiple independent overexpressing transformants of a putative rice XyG fucosyltransferase, O-acetyltransferase, and two galactosyltransferases were selected. Successful gene expression was confirmed by reverse transcription (RT)-PCR on homozygous T3 Arabidopsis transformants (Supplemental Fig. S8). The XyG structures in the transformants were assessed by XyG OLIMP and HPAEC. Expression of Os02g52630 (OsMUR2) in axy2.2 resulted in the restoration of fucosylated XyG to a level found in wild-type Arabidopsis plants, whereas the transformants of the other selected GT37 candidate genes showed no or only minor fucosylation, indicating that OsMUR2 is a true functional equivalent of AtMUR2 (Fig. 4; Supplemental Figs. S6–S9; Supplemental Table S3). Expression of the AXY4 ortholog Os06g12870 in the Arabidopsis axy4-3 mutant resulted in O-acetylated XyG but only to a limited extent (Fig. 4; Supplemental Figs. S4 and S10; Supplemental Table S4). These data indicate that Os06g12870, termed OsAXY4, exhibits XyG O-acetyltransferase activity but is not functionally equivalent to AtAXY4. The Arabidopsis double-mutant xlt2 mur3.1 contains an XyG that consists predominantly of XXXG (Jensen et al., 2012; Schultink et al., 2013). Expression of the Arabidopsis MUR3 homolog, Os03g05110, in this double mutant resulted in the generation of additional XyG oligosaccharides consistent by mass and retention time with XXLG and XXFG as well as their O-acetylated forms (Supplemental Figs. S5, S11, and S12; Supplemental Table S5), indicating that Os03g05110 is enzymatically equivalent to AtMUR3 and mediates the galactosylation of XXXG on the third xylosyl residue from the nonreducing end. Interestingly, most of these OsMUR3 transformants show a higher degree of galactosylation (81%–100%) compared with XyG found in Arabidopsis wild-type plants (68%), possibly because of the overexpression of OsMUR3 (Fig. 4; Supplemental Figs. S11 and S12). Expression of Os04g48480 in the Arabidopsis xlt2 mur3.1 double mutant resulted in the generation of additional oligosaccharides consistent by mass and retention time with XLXG (Fig. 4; Supplemental Figs. S11 and S12; Supplemental Table S5) but no additional fucosyl residues. This XyG structure resembles that of the Arabidopsis AtMUR3 single mutant, where XLT2 is still intact (Madson et al., 2003). Hence, Os04g48480 seems functionally equivalent to AtXLT2, which transfers galactosyl to the second xylosyl residue from the nonreducing end of XXXG. Therefore, Os03g05110 and Os04g48480 were named OsMUR3 and OsXLT2, respectively.
Figure 4.
XyG structures found in various Arabidopsis mutants and Arabidopsis transformants overexpressing particular rice genes. The XyG oligosaccharide mass profile is derived from 4-week-old leaves. The average relative ratios of fucosylated (A; XXFG, XXFG, XLFG, and XLFG), acetylated (B; XXLG, XXFG, and XLFG), and galactosylated (C and D; XXLG/XLXG, XXLG, XXFG, XXFG, XLLG, XLFG, and XLFG) oligosaccharides are shown. OX indicates an independent overexpression line. One-way ANOVA statistical analysis was performed. Different lowercase letters indicate significant differences. Error bars indicate sd (n ≥ 4).
Effects of Rice Candidate Genes on Arabidopsis Plant Morphology
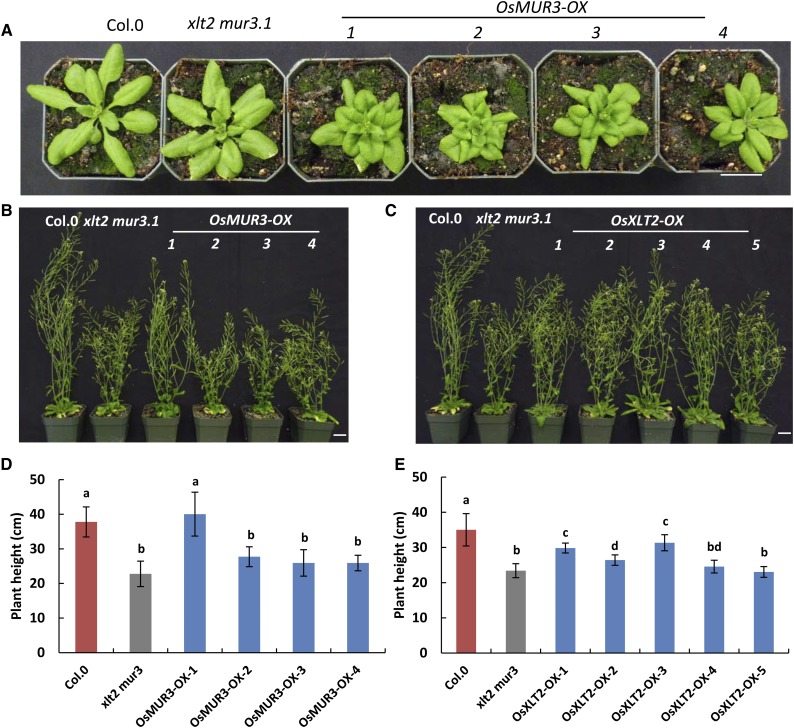
The Arabidopsis xlt2 mur3.1 double mutant displays dwarfism compared with wild-type or single-mutant plants (Jensen et al., 2012). To investigate whether complementation by OsMUR3 and/or OsXLT2 has an effect on dwarfism, the heights of T3-independent homozygous lines of the Arabidopsis transformants harboring OsXLT2 or OsMUR3 were measured and compared with Arabidopsis wild-type and xlt2 mur3.1 double-mutant plants. Overexpression of OsXLT2 in xlt2 mur3.1 only partially rescued the growth phenotype of mature plants (Fig. 5, C and E). Overexpression of OsMUR3 led to curly leaves, and only one of four independent OsMUR3 transformants displayed a rescued growth phenotype, whereas the other transformed lines remained dwarfed (Fig. 5, A, B, and D). Interestingly, the rescued line showed a low abundance of XyG galactosylation (81%) compared with other three lines (100%; Supplemental Figs. S11 and S12; Supplemental Table S5), suggesting that XyG overgalactosylation also affects plant growth. No significant plant or morphological differences between Arabidopsis wild-type and the axy4-3 and axy2.2/mur2 plants had been observed (Vanzin et al., 2002; Gille et al., 2011). Transforming these mutants with their corresponding rice orthologs also did not lead to any visible phenotypic alteration (Supplemental Figs. S13 and S14).
Figure 5.
Phenotype of rice galactosyltransferase transformants. Representative 4-week-old plants of OsMUR3 (A) and 7-week-old plants of OsMUR3 (B) or OsXLT2 (C) transformed into the xlt2 mur3.1 mutant. Bars = 2 cm. Average height of 7-week-old plants of OsMUR3 (D) and OsXLT2 (E) transformed into the xlt2 mur3.1 mutant. OX indicates an independent overexpression line. One-way ANOVA statistical analysis was performed. Different lowercase letters indicate significant differences. Error bars indicate sd (n = 12).
DISCUSSION
FucogalactoXyG Is Present in Rice
FucogalactoXyG has not been detected in moss (Physcomitrella patens), liverwort (Marchantia polymorpha), or green algae, such as members of the Charales, Coleochaetales, and Zygnematales, but is widely distributed in vascular plants from hornwort (Ceratophyllum submersum) to seed-bearing plants (Hoffman et al., 2005; Peña et al., 2008; Hsieh and Harris, 2009; Sørensen et al., 2011; Schultink et al., 2014). This led to the hypothesis that fucogalactoXyG appeared after the divergence of mosses and hornworts (Peña et al., 2008). Although fucogalactoXyG was thought to be absent in plant species of the Solanaceae, this type of XyG has recently been detected in tobacco pollen tubes (Lampugnani et al., 2013). FucogalactoXyG has not been found in plant tissues of the phylogenetically younger Poaceae. The hypothesis was raised that fucogalactoXyG has been potentially eliminated during evolution in these species. However, this study provides unambiguous evidence that fucogalactoXyG has been retained in species of the Poaceae, albeit only in specific tissues, such as the root epidermis, as well as tip-growing root hairs and possibly, pollen tubes.
This study, thus, shows that fucogalactoXyG can, indeed, be found in not only rice suspension cultured cells (Peña et al., 2008) but also, various tissues of the rice plant itself. The abundance of fucogalactoXyG is comparable with the XXGGn-type XyG in those tissues, indicating that this type of XyG is not just transiently present in these tissues. The main fucogalactoXyG oligosaccharides detected are XXFG but not XLFG, which has also been reported in walls of other plant families, such as members of the Juncaceae, Bromeliaceae, Cyperaceae, and Flagellariaceae in the Poales order other than the Poaceae (Hsieh and Harris, 2009). One possible hypothesis is that XLT2 galactosyltransferase orthologs are only lowly or not expressed in these species. Using an XyG-recognizing antibody, such as CCRC-M1, fucogalactoXyG was mainly localized only in the outer layer of rice root epidermal cells. Interestingly, the outer epidermis of Arabidopsis root also contains XyG with a higher degree of fucosylation (Freshour et al., 2003; Obel et al., 2009). This polarized localization of fucogalactoXyG suggests that this type of XyG may play an important role specifically at the interface of the plant root to the environment. FucogalactoXyG was also detected in rice root hairs, a tissue type where a growth phenotype has been observed in several Arabidopsis XyG mutants. These include the xyloglucan-specific galacturonosyltransferase1 (xut1) mutant as well as several xxt (XyG XXT) mutants. Particularly, the Arabidopsis mutant that lacks XyG in its walls, the xxt1 xxt2 double mutant (Cavalier et al., 2008), does not display any discernable growth or morphological phenotype in its aboveground tissues, but the root hairs are deformed or burst. Unexpectedly, only a few xxt double mutants were obtained when crossing the single xxt mutants (Cavalier et al., 2008), suggesting, perhaps, a defect in pollen transmission. Root hairs and pollen tubes represent tip-growing tissues. The retention of fucogalactoXyG in rice root tips and tobacco pollen suggests that this type of XyG may be essential for maintaining the functionality of these evolutionary conserved tissues. In contrast, in other tissues with extension growth in grasses and members of the Solanales, fucogalactoXyG was modified to an XXGGn-type XyG without apparent loss of function. In conclusion, the identification of fucosylated XyG in rice suggests that this type of XyG is evolutionary conserved in vascular plants, albeit that it is not the predominant type of XyG in grasses.
The Rice Genome Contains the Biosynthetic Genes to Generate FucogalactoXyG
AtMUR2 represents an XyG α-1,2 fucosyltransferase responsible for transferring a fucosyl residue from GDP-Fuc onto the galactosyl residue of the XyG side chain (Vanzin et al., 2002). Putative MUR2 orthologs were identified in rice based on amino acid sequence similarity, resulting in a distinctive clade among angiosperms and gymnosperms. Among these MUR2 orthologs, Os02g52640 has been previously identified as the only FUT1 ortholog in rice (Del Bem and Vincentz, 2010). However, our functional complementation study shows that overexpression of this gene in axy2.2 did not result in fucogalactoXyG, excluding XyG fucosyltransferase activity when expressed in Arabidopsis (Supplemental Fig. S9). In contrast, overexpressing the neighboring gene, Os02g52630, which was not placed in the XyG MUR2 clade in the previous study (Del Bem and Vincentz, 2010) but is coexpressed with OsCSLC3 in rice, resulted in fucogalactoXyG in axy2.2, indicating that this gene represents, indeed, a rice XyG fucosyltransferase. Whereas on the overall amino acid level, the identities of Os02g52630/OsMUR2 and Os02g52640640 with AtFUT1 are 47.62% and 42.63%, respectively, Os02g52640 lacks an N-terminal domain of approximately 200 amino acids that is present in AtMUR2/FUT1 and Os02g52630/OsMUR2 (Supplemental Fig. S15). It is, therefore, likely that this domain is required for XyG fucosyltransferase activity. It is possible that the neighboring Os02g52640 might have lost its enzymatic function during a recent gene duplication event from Os02g52630/OsMUR2.
Members of the TBL family have been shown to represent polysaccharide O-acetyltransferases (Gille et al., 2011; Xiong et al., 2013; Urbanowicz et al., 2014). XyG in the tbl27/axy4 mutant lacks acetyl substituents on the side chains of XyG. There are four rice homologs in the AtAXY4 subclade of the TBL family (Supplemental Fig. S4). Whereas AtAXY4 was able to completely restore XyG side-chain O-acetylation in the axy4 mutant (Gille et al., 2011), OsAXY4 leads to Arabidopsis XyG O-acetylation but to a much lesser extent. Because two of the other genes, Os07g15230 and Os07g15320, are closer to AtAXY4 in the phylogenic tree (Supplemental Fig. S4) and Os07g15320 is coexpressed with OsCSLC3 genes, it is possible that one or both genes might exhibit higher XyG acetyltransferase activity in Arabidopsis.
GT47 family members have been shown to be involved in XyG side-chain substitution, particularly galactosylation (Madson et al., 2003; Jensen et al., 2012; Peña et al., 2012). As shown here, OsXLT2 and OsMUR3 represent functional equivalents to their corresponding Arabidopsis orthologs when expressed in Arabidopsis. These genes are, thus, likely to play a role in the fucogalactoXyG synthesis in rice. Expression of OsMUR3 in Arabidopsis also resulted in unusual, hitherto unidentified structures (Supplemental Fig. S12, minor peak between XXXG and XXFG) that are not found in wild-type Arabidopsis plants. These data, thus, provide evidence that the substrate specificity of OsMUR3 might not be identical to AtMUR3. It is also not known if any of these genes also galactosylate the XXGGn-type XyG that is prevalent in rice. Reducing the degree of XyG galactosylation as observed in various mur3 mutant alleles or the xlt2 mur3.1 double mutant leads to dwarfed plants with curled rosette leaves (Jensen et al., 2012; Kong et al., 2015). When crossing such plants with XyG-depleted plants (xxt1 xxt2 double mutant), the phenotype is rescued, and the plants display a normal growth habit (Kong et al., 2015). Such data provide compelling evidence that plant growth is affected by the presence of an altered, dysfunctional XyG structure rather than the absence of XyG (Kong et al., 2015). Overexpressing tomato MUR3 (SlMUR3) and AtXLT2 in xlt2 mur3.1 and mur3-3 plants also leads to a rescue of the dwarfed growth phenotype of plants (Schultink et al., 2013; Kong et al., 2015). OsMUR3 overexpression as shown here was also able to rescue the growth phenotype of xlt2 mur3.1 in a single line (Fig. 5B). However, the shorter growth habit was retained in the other three lines, possibly caused by overexpression of OsMUR3 leading to XyG overgalactosylation, because no nongalactosylated XXXG was found in these transformants (Fig. 4C). Previous studies showed that overexpressing Eucalyptus grandis and tomato MUR3 in the Arabidopsis mur3 and xlt2 mur3.1 mutants also led to XyG overgalactosylation but not complete galactosylation. However, none of these plants were reported to exhibit a dwarfed phenotype (Lopes et al., 2010; Schultink et al., 2013). It, thus, seems that proper functionality of XyG is not retained when XyG galactosylation is below or above a certain threshold, which was found in the mur3 xlt2 double-mutant and the OsMUR3 overexpression lines or mur3-3 knockout plants, respectively. Overexpressing OsXLT2 in xlt2 mur3.1 resembled the XyG structural profile of the mur3.1 mutant plants, suggesting enzymatic equivalency to AtXLT2. Although the XyG structure is restored to mur3.1 levels, the growth phenotype remains stunted, although mur3.1 plants do not exhibit any growth defects, suggesting that OsXLT2 does not only affect XyG structure. It would be expected that those genes are expressed in the tissues, where the fucogalactoXyG is present in rice. Based on an online expression database, only OsMUR3 showed higher expression in pistil, emergence inflorescence, and embryo (Supplemental Fig. S16). Other genes, such as OsMUR2, are very lowly expressed and could not be detected by RT-PCR or real-time quantitative PCR (data not shown). Considering the polarized localization of fucogalactoXyG in root cells, it is likely that these genes have higher expression in those specific cells.
MATERIALS AND METHODS
Plant Material and Growth Conditions
The Arabidopsis (Arabidopsis thaliana) axy2.2 mutant is part of the XyG OLIMP screen collection (http://paulylab.berkeley.edu/axy-mutants.html). The axy2.2 mutant lacks fucosylated XyG as a cytosine deletion in the MUR2 gene (1,357 bp after start codon), which was found to cause premature translation termination. The Arabidopsis xlt2 mur3.1 double mutant was generated as previously described (Jensen et al., 2012). The axy4-3 mutant was obtained from the public Salk collection (Salk_044972; Gille et al., 2011). All seeds were stratified at 4°C for 1 to 2 d before sowing on soil. Soil-grown plants were grown under long-day conditions (16-h-light/8-h-dark cycle) with 70% humidity at 22°C. The transformant selection plates with seeds were placed in an environmentally controlled growth chamber under the same conditions as soil-grown plants for 2 weeks before transferring them to soil. All XyG structural analysis was performed on T3 homozygous lines, except for four negative fucosyltransferase transformants, for which positive T1 transformants were used. Seven-week-old plants were analyzed for their growth phenotype. Plant height was measured from the base of the stem to the most distal part of the plant. Rice (Oryza sativa) ‘Kitaake’ seeds were grown in an environmentally controlled chamber (12-h-light cycle at 28°C/12-h-dark cycle at 25°C, 55% humidity). Rice anthers were collected when the rice plants started flowering (approximately 10 weeks) and stored at −80°C. For rice young shoot and root material, rice seeds were sown on plates with two layers of filter paper soaked with 2 mL of water or one-half-strength Murashige and Skoog plates containing 1% (w/v) Suc. The plates were covered by two layers of aluminum foil and placed at 37°C for 3 d. Root and shoot tissues were separately collected and lyophilized or directly used for antibody labeling. Rice suspension cells were harvested by centrifugation at 4,000 rpm for 10 min; the pellet was washed four times with water and then lyophilized for additional analysis.
XyG Oligosaccharides Analysis
Alcohol-insoluble residue (AIR) was prepared as described (Günl et al., 2011). Briefly, rosette leaf material derived from 4-week-old Arabidopsis plants or rice tissues was collected and lyophilized. Dried material was ground in a Retsch Mill Grinder at 25 Hz for 150 s. The powder was extracted one time with 70% (v/v) aqueous ethanol, three times with chloroform:methanol (1:1), and one time with acetone and air dried, resulting in AIR material. The AIR material was incubated with 1 mL of 25 mm ammonium formate (pH 4.5) at 37°C overnight and centrifuged for 10 min at 14,000 rpm to remove buffer-soluble components. OLIMP of XyG was performed by digesting AIR with a purified xyloglucan-specific endoglucanase (XEG; no residual glycosidase or mixed-linkage glucanase activity) as described (Pauly et al., 1999b; Günl et al., 2010; Jensen et al., 2012). The spectra of the solubilized XyG oligosaccharides were obtained using an Axima matrix-assisted laser desorption/ionization time-of-flight instrument (Shimadzu) set to linear positive mode with an acceleration voltage of 20,000 V. For separation of XyG oligosaccharides by high-performance anion-exchange chromatography with pulsed amperometric detection (HPAEC-PAD), XEG-released XyG oligosaccharides were subjected to a CarboPac PA200 Column. The XyG oligosaccharides were separated using a linear gradient of 0 to 80 mm sodium acetate in 100 mm sodium hydroxide within 15 min at a flow rate of 0.4 mL min−1. To identify XyG oligosaccharides in HPAEC-PAD peaks, drops eluted from 10 to 18 min were collected separately. Around 10 µL of collected samples was desalted with ion-exchange resin (Bio-Rad), and 10 ng of xylohexoase was added as an internal control (Megazyme). Around 3 µL of desalted samples was used for XyG oligosaccharides spectra collection by the matrix-assisted laser desorption/ionization time-of-flight instrument as mentioned above.
Glycosidic Linkage Analysis
Linkage analysis was performed according to the previously described method by generating partially methylated alditol acetate (PMAA) derivatives (Cavalier et al., 2008). For this purpose, roughly 2 mg of AIR was digested with XEG overnight at 37°C. The supernatant containing the released XyG oligosaccharides was derivatized. Methylated samples were hydrolyzed, reduced with NaBD4 (Sigma-Aldrich), and per O-acetylated. The PMAA derivatives were analyzed on an Agilent 6890 Series GC System equipped with a 5975B Inert XL MSD and an SP-2380 Fused Silica Capillary Column (30-m × 0.25-mm i.d. × 20-μm film thickness; Supelco). Glycosydic linkages were assigned based on the retention times of known standards (H-Trisaccharide from Sigma-Aldrich for the detection of T-Fucp and 2-Galp and XyG oligosaccharides from Megazyme for the detection of other XyG glycosydic linkages) and mass spectra in the PMAA database (http://www.ccrc.uga.edu/specdb/ms/pmaa/pframe.html). The retention times of 2-Galp and 6-Glcp are so close that the two peaks overlapped. The ion with m/z of 205, which is specific to 2-Galp, was used to quantify the amount of 2-Galp.
Antibody Labeling
Rice shoot and root (3 d old) were fixed overnight in 4% (v/v) formaldehyde phosphate-buffered saline (137 mm sodium chloride, 2.7 mm potassium chloride, 8.1 mm disodium hydrogen phosphate, 1.76 mm potassium dihydrogen phosphate, pH 7.4) at 4°C. After fixation, plant material was rinsed with Tris-buffered saline (TBS; 50 mm Tris-Cl, pH 7.6, 150 mm NaCl, pH 7.5) and used for hand sections. Sections were placed in 2-mL Eppendorf tubes with 1% (w/v) bovine serum albumin in Tris-buffered saline (BTBS) for blocking. Sections were then labeled with the primary antibodies CCRC-M1, CCRC-48 (glycomics.ccrc.uga.edu; mouse monoclonal; Puhlmann et al., 1994), or LM11 (www.plantprobes.net/index.php; rat monoclonal; McCartney et al., 2005) by overnight incubation under rotating conditions in the cold room with a 1:50 (v:v) antibody dilution in BTBS. Sections were rinsed three times in TBS and incubated at room temperature with rotation for 2 h with fluorescein isothiocyanate (FITC)-conjugated secondary antibodies (Sigma-Aldrich) in 1:100 (v:v) dilutions in BTBS. Anti-mouse IgG (whole molecule)-FITC and anti-rat IgG (whole molecule)-FITC antibody were used as secondary antibodies for CCRC-M1 and CCRC-M48 and LM11, respectively. Sections were washed three times with TBS to remove any excess secondary antibody and then transferred to microscope slides. Fluorescence was observed with a Leica DFC 300FX Epifluorescence Microscope (Leica).
Bioinformatics Analysis of Coexpressed XyG Biosynthetic Genes and MUR2, XLT2, MUR3, and AXY4 Homologs
The coexpression experiment was performed using GeneCAT (Mutwil et al., 2008). Affymetrix probe set OsAffx.17009.1.S1_x_at was used as the query for coexpression genes with a cutoff at 0.7 (genecat.mpg.de/). Only the top 50 coexpressed genes are listed in Supplemental Table S2. The genes with annotation putative involved in XyG synthesis were picked and listed in Table II. The Arabidopsis MUR2/FUT1, MUR3, XLT2, and AXY4/TBL27 protein sequences were obtained from The Arabidopsis Information Resource (www.arabidopsis.org). BLAST (Altschul et al., 1997) searches were performed using these protein sequences to identify homologs in Phytozome (www.phytozome.net). Multiple sequence alignment was performed using ClustalX2 (Larkin et al., 2007), and a neighbor-joining tree was constructed using Mega5 with default parameters (Tamura et al., 2011).
Cloning and Transformation of Rice OsMUR2, OsXLT2, OsMUR3, and OsAXY4 into Arabidopsis Mutants
The complementary DNA (cDNA) of OsAXY4, OsXLT2, OsMUR3, OsMUR2, and another four fucosyltransferase candidates was ordered from the Rice Genome Resource Center (www.rgrc.dna.affrc.go.jp). The coding sequence of these genes was amplified using the primers listed in Supplemental Table S6 with the Fusion enzyme (New England Biolabs). The PCR products were cloned into the pZErO vector (Invitrogen) with a BP reaction and subcloned into the overexpression plasmid pORE E4 containing a tobacco (Nicotiana alata) Cryptic Constitutive Promoter2 promoter (Coutu et al., 2007) or pGWB511 with a 35S promoter (for OsAXY4; Nakagawa et al., 2009) with an LR reaction. The plasmids were transformed into the Agrobacterium tumefaciens strain GV3101. Positive colonies were selected and subsequently transformed into the corresponding Arabidopsis mutant plants using the floral dip method (Clough and Bent, 1998). Positive transformants were identified by selection on one-half-strength Murashige and Skoog and 1% (w/v) Suc with 50-µg mL−1 kanamycin plates (OsMUR2, OsXLT2, and OsMUR3 transformants) or 50-µg mL−1 hygromycin plates (OsAXY4 transformant) in the T1 generation and genotyped to confirm a stable transformation and correct mutant background in the T1 and T2 generations using the primers listed in Supplemental Table S6. Homozygous transformants were identified based on segregation of kanamycin or hygromycin resistance in the T3 generation.
RT-PCR
Four-week-old Arabidopsis leaf material was harvested for transcript analysis. RNA was extracted with an RNeasy Plant Mini Kit (Qiagen) according to the manual. DNA was removed from the RNA samples by treatment with DNaseI (Roche Applied Science). cDNA synthesis was performed according to the manual using the SuperScript III First-Strand Synthesis SuperMix Kit (Life Technologies) with oligo(dT) primers. The newly synthesized cDNA was diluted to 80 μL total, and its concentration was measured. RT-PCR was performed with JumpStart Red-taq Ready Mix (Sigma-Aldrich) using the primers listed in Supplemental Table S6.
Rice gene sequence data from this article can be found in the Rice Genome Annotation Project (rice.plantbiology.msu.edu), Knowledge-based Oryza Molecular Biological Encyclopedia (cdna01.dna.affrc.go.jp/cDNA/), and Rice Genome Resource Center (www.rgrc.dna.affrc.go.jp) databases under accession numbers OsMUR2, LOC_Os02g52630 and AK102499; OsMUR3, LOC_Os03g05110 and AK103041; OsXLT2, LOC_Os04g48480 and AK069178; and OsAXY4, LOC_Os06g12870 and AK240937. Arabidopsis gene sequence data from this article can be found in The Arabidopsis Information Resource under accession numbers MUR2/FUT1, AT2G03220; MUR3, AT2G20370; XLT2, AT5G62220; AXY4, AT1G70230; and Polypyrimidine Tract-Binding Protein, AT3G01150.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. HPAEC-PAD analysis of XyG derived from different tissues.
Supplemental Figure S2. Immunofluorescent labeling of rice 3-d-old root and shoot transverse sections with different antibodies.
Supplemental Figure S3. Phylogenic tree of MUR2 homologs in Arabidopsis, rice, and Brachypodium.
Supplemental Figure S4. Phylogenic tree of AXY4 homologs in Arabidopsis, rice, and Brachypodium.
Supplemental Figure S5. Phylogenic tree of MUR3 and XLT2 homologs in Arabidopsis, rice, and Brachypodium.
Supplemental Figure S6. XyG OLIMP of 4-week-old leaves from Arabidopsis Col-0, axy2.2/fut1, and OsMUR2 transformants into the axy2.2 mutant.
Supplemental Figure S7. XyG HPAEC spectra derived from cell walls of 4-week-old leaves from Col-0, axy2.2, and OsMUR2 transformants into the axy2.2 mutant.
Supplemental Figure S8. RT-PCR analysis of total RNA extracted from 4-week-old leaves of Arabidopsis wild types (Columbia-0 [Col-0]), mutants, and all transformants (OsMUR2-OX-axy2.2, OsAXY4-OX-axy4-3, OsXLT2-OX-xlt2 mur3, and OsMUR3-OX-xlt2 mur3).
Supplemental Figure S9. XyG OLIMP of 4-week-old leaves from Arabidopsis Col-0 and rice homologs of AtMUR2 transformants into the axy2.2 mutant.
Supplemental Figure S10. XyG OLIMP of 4-week-old leaves from Arabidopsis Col-0, axy4-3, and OsAXY4 transformants into the axy4-3 mutant.
Supplemental Figure S11. XyG OLIMP of 4-week-old leaves from Arabidopsis Col-0, xlt2 mur3.1, OsMUR3, and OsXLT2 transformants into the xlt2 mur3 mutant.
Supplemental Figure S12. XyG HPAEC spectra derived from cell walls of 4-week-old leaves from Col-0, xlt2 mur3.1, OsMUR3, and OsXLT2 transformants into the xlt2 mur3 mutant.
Supplemental Figure S13. Phenotype of 7-week-old plants of Col-0, axy2.2, and OsMUR2 transformants into the axy2.2 mutant.
Supplemental Figure S14. Phenotype of 7-week-old plants of Col-0, axy4-3, and OsAXY4 transformants into the axy4-3 mutant.
Supplemental Figure S15. Sequence alignment of AtFUT1/MUR2 with Os02g52630/OsMUR2 and Os02g52640.
Supplemental Figure S16. RNA sequencing fragments per kilobase of exon per million fragments mapped expression values of rice genes in different tissues.
Supplemental Table S1. List of m/z values with known or predicted XyG structures shown in this study.
Supplemental Table S2. List of the top 50 coexpressed genes with OsCSLC3.
Supplemental Table S3. XyG oligosaccharide profile of Col-0, axy2.2, and OsMUR2 transformants into the axy2.2 mutant by mass profiling.
Supplemental Table S4. XyG oligosaccharides profile of Col-0, axy4-3, and OsAXY4 transformants into the axy4-3 mutant by mass profiling.
Supplemental Table S5. XyG oligosaccharides profile of Col-0, xlt2 mur3.1, OsMUR3, and OsXLT2 transformants into the xlt2 mur3.1 mutant by mass profiling.
Supplemental Table S6. Primers used in this study.
Supplementary Material
Acknowledgments
We thank Kirk Schnorr (Novozymes, Bagsvaerd, Denmark) for the XEG; Henrik Scheller (Joint Bioenergy Institute) for the LM11 antibody; and Pam Ronalds (University of California, Davis), Joshua Heazlewood (Joint Bioenergy Institute), Minguel Vega-Sanchez (Joint Bioenergy Institute), and Susana Gonzalez Fernandez-Nino (Joint Bioenergy Institute) for providing the rice suspension cells and rice seeds.
Glossary
- AIR
alcohol-insoluble residue
- BTBS
bovine serum albumin in Tris-buffered saline
- cDNA
complementary DNA
- Col-0
Columbia-0
- FITC
fluorescein isothiocyanate
- HPAEC
high-performance anion-exchange chromatography
- HPAEC-PAD
high-performance anion-exchange chromatography with pulsed amperometric detection
- m/z
mass to charge ratio
- OLIMP
oligosaccharide mass profiling
- PMAA
partially methylated alditol acetate
- RT
reverse transcription
- TBS
Tris-buffered saline
- XEG
xyloglucan-specific endoglucanase
- XyG
xyloglucan
Footnotes
This work was supported by the Energy Bioscience Institute.
Articles can be viewed without a subscription.
References
- Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25: 3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bischoff V, Nita S, Neumetzler L, Schindelasch D, Urbain A, Eshed R, Persson S, Delmer D, Scheible WR (2010) TRICHOME BIREFRINGENCE and its homolog AT5G01360 encode plant-specific DUF231 proteins required for cellulose biosynthesis in Arabidopsis. Plant Physiol 153: 590–602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bootten TJ, Harris PJ, Melton LD, Newman RH (2004) Solid-state 13C-NMR spectroscopy shows that the xyloglucans in the primary cell walls of mung bean (Vigna radiata L.) occur in different domains: a new model for xyloglucan-cellulose interactions in the cell wall. J Exp Bot 55: 571–583 [DOI] [PubMed] [Google Scholar]
- Brennan M, Harris PJ (2011) Distribution of fucosylated xyloglucans among the walls of different cell types in monocotyledons determined by immunofluorescence microscopy. Mol Plant 4: 144–156 [DOI] [PubMed] [Google Scholar]
- Carpita NC, Gibeaut DM (1993) Structural models of primary cell walls in flowering plants: consistency of molecular structure with the physical properties of the walls during growth. Plant J 3: 1–30 [DOI] [PubMed] [Google Scholar]
- Cavalier DM, Lerouxel O, Neumetzler L, Yamauchi K, Reinecke A, Freshour G, Zabotina OA, Hahn MG, Burgert I, Pauly M, et al. (2008) Disrupting two Arabidopsis thaliana xylosyltransferase genes results in plants deficient in xyloglucan, a major primary cell wall component. Plant Cell 20: 1519–1537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Cocuron JC, Lerouxel O, Drakakaki G, Alonso AP, Liepman AH, Keegstra K, Raikhel N, Wilkerson CG (2007) A gene from the cellulose synthase-like C family encodes a β-1,4 glucan synthase. Proc Natl Acad Sci USA 104: 8550–8555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosgrove DJ. (2005) Growth of the plant cell wall. Nat Rev Mol Cell Biol 6: 850–861 [DOI] [PubMed] [Google Scholar]
- Coutu C, Brandle J, Brown D, Brown K, Miki B, Simmonds J, Hegedus DD (2007) pORE: a modular binary vector series suited for both monocot and dicot plant transformation. Transgenic Res 16: 771–781 [DOI] [PubMed] [Google Scholar]
- Dardelle F, Le Mauff F, Lehner A, Loutelier-Bourhis C, Bardor M, Rihouey C, Causse M, Lerouge P, Driouich A, Mollet JC (2015) Pollen tube cell walls of wild and domesticated tomatoes contain arabinosylated and fucosylated xyloglucan. Ann Bot (Lond) 115: 55–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Del Bem LE, Vincentz MG (2010) Evolution of xyloglucan-related genes in green plants. BMC Evol Biol 10: 341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faik A, Price NJ, Raikhel NV, Keegstra K (2002) An Arabidopsis gene encoding an α-xylosyltransferase involved in xyloglucan biosynthesis. Proc Natl Acad Sci USA 99: 7797–7802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freshour G, Bonin CP, Reiter WD, Albersheim P, Darvill AG, Hahn MG (2003) Distribution of fucose-containing xyloglucans in cell walls of the mur1 mutant of Arabidopsis. Plant Physiol 131: 1602–1612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fry SC, York WS, Albersheim P, Darvill A, Hayashi T, Joseleau JP, Kato Y, Lorences EP, Maclachlan GA, McNeil M, et al. (1993) An unambiguous nomenclature for xyloglucan-derived oligosaccharides. Physiol Plant 89: 1–3 [Google Scholar]
- Gibeaut DM, Pauly M, Bacic A, Fincher GB (2005) Changes in cell wall polysaccharides in developing barley (Hordeum vulgare) coleoptiles. Planta 221: 729–738 [DOI] [PubMed] [Google Scholar]
- Gille S, de Souza A, Xiong G, Benz M, Cheng K, Schultink A, Reca IB, Pauly M (2011) O-acetylation of Arabidopsis hemicellulose xyloglucan requires AXY4 or AXY4L, proteins with a TBL and DUF231 domain. Plant Cell 23: 4041–4053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gille S, Pauly M (2012) O-acetylation of plant cell wall polysaccharides. Front Plant Sci 3: 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Günl M, Gille S, Pauly M (2010) OLIgo mass profiling (OLIMP) of extracellular polysaccharides. J Vis Exp 40: e2046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Günl M, Kraemer F, Pauly M (2011) Oligosaccharide mass profiling (OLIMP) of cell wall polysaccharides by MALDI-TOF/MS. Methods Mol Biol 715: 43–54 [DOI] [PubMed] [Google Scholar]
- Hayashi T, Kaida R (2011) Functions of xyloglucan in plant cells. Mol Plant 4: 17–24 [DOI] [PubMed] [Google Scholar]
- Hoffman M, Jia Z, Peña MJ, Cash M, Harper A, Blackburn AR II, Darvill A, York WS (2005) Structural analysis of xyloglucans in the primary cell walls of plants in the subclass Asteridae. Carbohydr Res 340: 1826–1840 [DOI] [PubMed] [Google Scholar]
- Hsieh YS, Harris PJ (2009) Xyloglucans of monocotyledons have diverse structures. Mol Plant 2: 943–965 [DOI] [PubMed] [Google Scholar]
- Hsieh YS, Harris PJ (2012) Structures of xyloglucans in primary cell walls of gymnosperms, monilophytes (ferns sensu lato) and lycophytes. Phytochemistry 79: 87–101 [DOI] [PubMed] [Google Scholar]
- Jensen JK, Schultink A, Keegstra K, Wilkerson CG, Pauly M (2012) RNA-Seq analysis of developing nasturtium seeds (Tropaeolum majus): identification and characterization of an additional galactosyltransferase involved in xyloglucan biosynthesis. Mol Plant 5: 984–992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia Z, Cash M, Darvill AG, York WS (2005) NMR characterization of endogenously O-acetylated oligosaccharides isolated from tomato (Lycopersicon esculentum) xyloglucan. Carbohydr Res 340: 1818–1825 [DOI] [PubMed] [Google Scholar]
- Jia Z, Qin Q, Darvill AG, York WS (2003) Structure of the xyloglucan produced by suspension-cultured tomato cells. Carbohydr Res 338: 1197–1208 [DOI] [PubMed] [Google Scholar]
- Kato Y, Ito S, Iki K, Matsuda K (1982) Xyloglucan and rβ-D-glucan in cell walls of rice seedlings. Plant Cell Physiol 23: 351–364 [Google Scholar]
- Kato Y, Ito S, Mitsuishi Y (2004) Further structural study of the barley and bamboo shoot xyloglucans by xyloglucanase. J Appl Glycosci 51: 327–333 [Google Scholar]
- Kong Y, Peña MJ, Renna L, Avci U, Pattathil S, Tuomivaara ST, Li X, Reiter WD, Brandizzi F, Hahn MG, et al. (2015) Galactose-depleted xyloglucan is dysfunctional and leads to dwarfism in Arabidopsis. Plant Physiol 167: 1296–1306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lampugnani ER, Moller IE, Cassin A, Jones DF, Koh PL, Ratnayake S, Beahan CT, Wilson SM, Bacic A, Newbigin E (2013) In vitro grown pollen tubes of Nicotiana alata actively synthesise a fucosylated xyloglucan. PLoS ONE 8: e77140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, et al. (2007) Clustal W and Clustal X version 2.0. Bioinformatics 23: 2947–2948 [DOI] [PubMed] [Google Scholar]
- Li X, Cordero I, Caplan J, Mølhøj M, Reiter WD (2004) Molecular analysis of 10 coding regions from Arabidopsis that are homologous to the MUR3 xyloglucan galactosyltransferase. Plant Physiol 134: 940–950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopes F, Pauly M, Brommonshenkel S, Lau E, Diola V, Passos J, Loureiro M (2010) The EgMUR3 xyloglucan galactosyltransferase from Eucalyptus grandis complements the mur3 cell wall phenotype in Arabidopsis thaliana. Tree Genet Genomes 6: 745–756 [Google Scholar]
- Madson M, Dunand C, Li X, Verma R, Vanzin GF, Caplan J, Shoue DA, Carpita NC, Reiter WD (2003) The MUR3 gene of Arabidopsis encodes a xyloglucan galactosyltransferase that is evolutionarily related to animal exostosins. Plant Cell 15: 1662–1670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansoori N, Schultink A, Schubert J, Pauly M (2015) Expression of heterologous xyloglucan xylosyltransferases in Arabidopsis to investigate their role in determining xyloglucan xylosylation substitution patterns. Planta 241: 1145–1158 [DOI] [PubMed] [Google Scholar]
- McCartney L, Marcus SE, Knox JP (2005) Monoclonal antibodies to plant cell wall xylans and arabinoxylans. J Histochem Cytochem 53: 543–546 [DOI] [PubMed] [Google Scholar]
- McDougall GJ, Fry SC (1994) Fucosylated xyloglucan in suspension-cultured cells of the graminaceous monocotyledon, Festuca arundinacea. Plant Physiol 143: 591–595 [Google Scholar]
- Mutwil M, Obro J, Willats WG, Persson S (2008) GeneCAT—novel webtools that combine BLAST and co-expression analyses. Nucleic Acids Res 36: W320–W326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa T, Ishiguro S, Kimura T (2009) Gateway vectors for plant transformation. Plant Biotechnol 26: 275–284 [Google Scholar]
- Obel N, Erben V, Schwarz T, Kühnel S, Fodor A, Pauly M (2009) Microanalysis of plant cell wall polysaccharides. Mol Plant 2: 922–932 [DOI] [PubMed] [Google Scholar]
- Park YB, Cosgrove DJ (2012) A revised architecture of primary cell walls based on biomechanical changes induced by substrate-specific endoglucanases. Plant Physiol 158: 1933–1943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauly M, Albersheim P, Darvill A, York WS (1999a) Molecular domains of the cellulose/xyloglucan network in the cell walls of higher plants. Plant J 20: 629–639 [DOI] [PubMed] [Google Scholar]
- Pauly M, Andersen LN, Kauppinen S, Kofod LV, York WS, Albersheim P, Darvill A (1999b) A xyloglucan-specific endo-β-1,4-glucanase from Aspergillus aculeatus: expression cloning in yeast, purification and characterization of the recombinant enzyme. Glycobiology 9: 93–100 [DOI] [PubMed] [Google Scholar]
- Pauly M, Gille S, Liu L, Mansoori N, de Souza A, Schultink A, Xiong G (2013) Hemicellulose biosynthesis. Planta 238: 627–642 [DOI] [PubMed] [Google Scholar]
- Pauly M, Qin Q, Greene H, Albersheim P, Darvill A, York WS (2001) Changes in the structure of xyloglucan during cell elongation. Planta 212: 842–850 [DOI] [PubMed] [Google Scholar]
- Peña MJ, Darvill AG, Eberhard S, York WS, O’Neill MA (2008) Moss and liverwort xyloglucans contain galacturonic acid and are structurally distinct from the xyloglucans synthesized by hornworts and vascular plants. Glycobiology 18: 891–904 [DOI] [PubMed] [Google Scholar]
- Peña MJ, Kong Y, York WS, O’Neill MA (2012) A galacturonic acid-containing xyloglucan is involved in Arabidopsis root hair tip growth. Plant Cell 24: 4511–4524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrin RM, DeRocher AE, Bar-Peled M, Zeng W, Norambuena L, Orellana A, Raikhel NV, Keegstra K (1999) Xyloglucan fucosyltransferase, an enzyme involved in plant cell wall biosynthesis. Science 284: 1976–1979 [DOI] [PubMed] [Google Scholar]
- Puhlmann J, Bucheli E, Swain MJ, Dunning N, Albersheim P, Darvill AG, Hahn MG (1994) Generation of monoclonal antibodies against plant cell-wall polysaccharides; I. Characterization of a monoclonal antibody to a terminal α-(1—>2)-linked fucosyl-containing epitope. Plant Physiol 104: 699–710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultink A, Cheng K, Park YB, Cosgrove DJ, Pauly M (2013) The identification of two arabinosyltransferases from tomato reveals functional equivalency of xyloglucan side chain substituents. Plant Physiol 163: 86–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultink A, Liu L, Zhu L, Pauly M (2014) Structural diversity and function of xyloglucan sidechain substituents. Plants 3: 526–542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sørensen I, Pettolino FA, Bacic A, Ralph J, Lu F, O’Neill MA, Fei Z, Rose JK, Domozych DS, Willats WG (2011) The charophycean green algae provide insights into the early origins of plant cell walls. Plant J 68: 201–211 [DOI] [PubMed] [Google Scholar]
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28: 2731–2739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuomivaara ST, Yaoi K, O’Neill MA, York WS (2015) Generation and structural validation of a library of diverse xyloglucan-derived oligosaccharides, including an update on xyloglucan nomenclature. Carbohydr Res 402: 56–66 [DOI] [PubMed] [Google Scholar]
- Urbanowicz BR, Peña MJ, Moniz HA, Moremen KW, York WS (2014) Two Arabidopsis proteins synthesize acetylated xylan in vitro. Plant J 80: 197–206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanzin GF, Madson M, Carpita NC, Raikhel NV, Keegstra K, Reiter WD (2002) The mur2 mutant of Arabidopsis thaliana lacks fucosylated xyloglucan because of a lesion in fucosyltransferase AtFUT1. Proc Natl Acad Sci USA 99: 3340–3345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vuttipongchaikij S, Brocklehurst D, Steele-King C, Ashford DA, Gomez LD, McQueen-Mason SJ (2012) Arabidopsis GT34 family contains five xyloglucan α-1,6-xylosyltransferases. New Phytol 195: 585–595 [DOI] [PubMed] [Google Scholar]
- Watanabe T, Shida M, Murayama T, Furuyama Y, Nakajima T, Matsuda K, Kainuma K (1984) Xyloglucan in cell walls of rice hull. Carbohydr Res 129: 229–242 [Google Scholar]
- Xiong G, Cheng K, Pauly M (2013) Xylan O-acetylation impacts xylem development and enzymatic recalcitrance as indicated by the Arabidopsis mutant tbl29. Mol Plant 6: 1373–1375 [DOI] [PubMed] [Google Scholar]
- York WS, Kumar Kolli VS, Orlando R, Albersheim P, Darvill AG (1996) The structures of arabinoxyloglucans produced by solanaceous plants. Carbohydr Res 285: 99–128 [DOI] [PubMed] [Google Scholar]
- Zabotina OA, van de Ven WT, Freshour G, Drakakaki G, Cavalier D, Mouille G, Hahn MG, Keegstra K, Raikhel NV (2008) Arabidopsis XXT5 gene encodes a putative α-1,6-xylosyltransferase that is involved in xyloglucan biosynthesis. Plant J 56: 101–115 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.