CENP-32 depletion releases centrosomes from spindles after initiating spindle assembly. The free centrosomes do not interfere with the structure or function of the bipolar anastral spindle. The asters appear to be able to interact with the surface of the spindle but are unable to incorporate into it.
Abstract
Centrosomes nucleate spindle formation, direct spindle pole positioning, and are important for proper chromosome segregation during mitosis in most animal cells. We previously reported that centromere protein 32 (CENP-32) is required for centrosome association with spindle poles during metaphase. In this study, we show that CENP-32 depletion seems to release centrosomes from bipolar spindles whose assembly they had previously initiated. Remarkably, the resulting anastral spindles function normally, aligning the chromosomes to a metaphase plate and entering anaphase without detectable interference from the free centrosomes, which appear to behave as free asters in these cells. The free asters, which contain reduced but significant levels of CDK5RAP2, show weak interactions with spindle microtubules but do not seem to make productive attachments to kinetochores. Thus CENP-32 appears to be required for centrosomes to integrate into a fully functional spindle that not only nucleates astral microtubules, but also is able to nucleate and bind to kinetochore and central spindle microtubules. Additional data suggest that NuMA tethers microtubules at the anastral spindle poles and that augmin is required for centrosome detachment after CENP-32 depletion, possibly due to an imbalance of forces within the spindle.
INTRODUCTION
Accurate spindle formation is essential for the proper segregation of chromosomes during cell division. The bipolar mitotic spindle is composed of polar microtubules organized with their minus ends anchored to centrosomes and plus ends projecting outward to form the spindle and asters. In Saccharomyces cerevisiae, the spindle pole body, which is functionally equivalent to the centrosome, serves as the microtubule-organizing center and contributes to bipolar spindle formation in the mitotic nucleus (Byers and Goetsch, 1974).
In animal cells, spindles usually form as the result of the separation and activation of centrosomes during the G2 phase of the cell cycle (Kimura et al., 1997; Wakefield et al., 2000; Jackman et al., 2003; Doxsey et al., 2005; Hachet et al., 2007; Portier et al., 2007).
The pericentriolar material (PCM) is created by pericentrin/kendrin and centrosome and Golgi–localized, PKN-associated protein (CG-NAP), which are stabilized by Kizuna (Oshimori et al., 2006, 2009). CG-NAP and kendrin help to organize microtubule nucleation in the mammalian centrosome by anchoring the γ-tubulin ring complex (γ-TuRC) together with several other proteins (Dictenberg et al., 1998; Hung et al., 2000; Takahashi et al., 2000, 2002; Casenghi et al., 2003). Centrosomal localization of CG-NAP and Kizuna is regulated by Cep72, which is also involved in γTuRC recruitment to the centrosome (Oshimori et al., 2009).
Centrosomes nucleate the assembly of highly dynamic microtubules that shift between states of polymerization and depolymerization, effectively probing the cytoplasm and enabling them to be captured by kinetochores on mitotic chromosomes (Mitchison and Kirschner, 1985; Kirschner and Mitchison, 1986). A subset of microtubule plus ends from opposite poles also takes part in antiparallel interactions in the midzone and stabilizes the central spindle. Correct centrosome positioning and microtubule nucleation in the mitotic cell are thus extremely important for establishing and maintaining normal spindle shape and structure (O'Connell and Wang, 2000; Lüders and Stearns, 2007; Dunsch et al., 2012; Laan et al., 2012; Kiyomitsu and Cheeseman, 2013).
Despite the obvious importance of centrosomes in nucleating formation of bipolar spindles, spindles can form in the absence of centrosomes (e.g., in oocytes) and after centrosome inactivation (Uzbekov et al., 1995; Khodjakov et al., 2000). Centrosome-independent mechanisms for mitotic spindle formation appear to involve the Ran-GTP–mediated polymerization of microtubules near chromosomes (Karsenti et al., 1984; Heald et al., 1996; Wilde and Zheng, 1999; Wilde et al., 2001; Kalab et al., 2002; O'Connell and Khodjakov, 2007), capture of the microtubule plus ends by kinetochores, and clustering of the microtubule minus ends by microtubule motor proteins to form the spindle poles (Sawin et al., 1992; Merdes et al., 1996; Walczak et al., 1998; Sharp et al., 2000). Ran is also required for the correct cortical localization of nuclear mitotic apparatus protein (NuMA; Wilde and Zheng, 1999; Zhang et al., 1999; Wiese et al., 2001). NuMA interacts with cytoplasmic dynein and dynactin, and they transport it toward microtubule minus ends (Merdes et al., 1996, 2000). These motor proteins accumulate at the pericentrosomal region and play a role in generating pulling forces on astral microtubules (Gaglio et al., 1995; Haren et al., 2009).
De novo microtubule generation can occur within mitotic spindles of vertebrate cells in a centrosome-independent manner (Goshima et al., 2008). The dim γ-tubulin 2-9 (Dgt2-9) octamer complex, or augmin, was reported to nucleate microtubule formation within mitotic spindles in a γ-tubulin–dependent process (Uehara et al., 2009). Thus there may exist more than one centrosome-independent spindle formation mechanism that maintains acentrosomal spindles in mitosis across a wide range of species, including plants (Masoud et al., 2013).
Previously our mitotic chromosome proteomics analysis predicted mitotic chromosome association of centromere protein 32 (CENP-32)/C9orf119. Streptavidin-binding peptide–tagged CENP-32 accumulated on mitotic spindles and at kinetochores (Supplemental Figure S1A; Ohta et al., 2010). Our proteomics-based bioinformatics MCCP (multi-classifier combinatorial proteomics) analysis predicted that CENP-32 function is related to CLASP1 and 2, whose association with microtubules, kinetochores, and centrosomes is well known. CLASPs are involved in multiple microtubule-dependent processes in mitosis, including kinetochore fiber dynamics, mitotic spindle assembly, and the stabilization of microtubule plus ends near the cortex (Maiato et al., 2005; Mimori-Kiyosue et al., 2005; Miller et al., 2009; Reis et al., 2009; Patel et al., 2012; Bird et al., 2013). Recent work identified CLASP1 as being required for proper positioning of the mitotic spindle (Samora et al., 2011; Espiritu et al., 2012; Kiyomitsu and Cheeseman, 2012).
We previously reported that centrosomes detach from mitotic spindles in CENP-32–depleted cells (Ohta et al., 2010). A second pericentriolar protein, CDK5RAP2, is also required to maintain centriole engagement and cohesion (Fong et al., 2008; Barrera et al., 2010). CDK5RAP2 was reported to be recruited to centrosomes by dynein and be required to maintain attachment of centrosomes to spindle poles in mitosis (Barr et al., 2010; Jia et al., 2013).
It is unclear how the link between centrosomes and the mitotic spindle poles is maintained. Here we study whether CENP-32–depleted spindles, which apparently do not form a stable connection with centrosomes, could facilitate normal chromosome arrangements to a metaphase plate and segregation during anaphase. We find that CENP-32 is necessary for CG-NAP recruitment to centrosomes and for maintaining interactions between the mitotic spindle and centrosomes. Moreover, augmin is necessary for the centrosome detachment and subsequent anastral mitosis that occurs in the absence of CENP-32. Our findings demonstrate that CENP-32 is required for centrosome integration into the spindle and that its absence causes abnormal microtubule organization, possibly by interfering with CG-NAP localization and/or function.
RESULTS
CENP-32 depletion leads to centrosome mislocalization from spindle poles
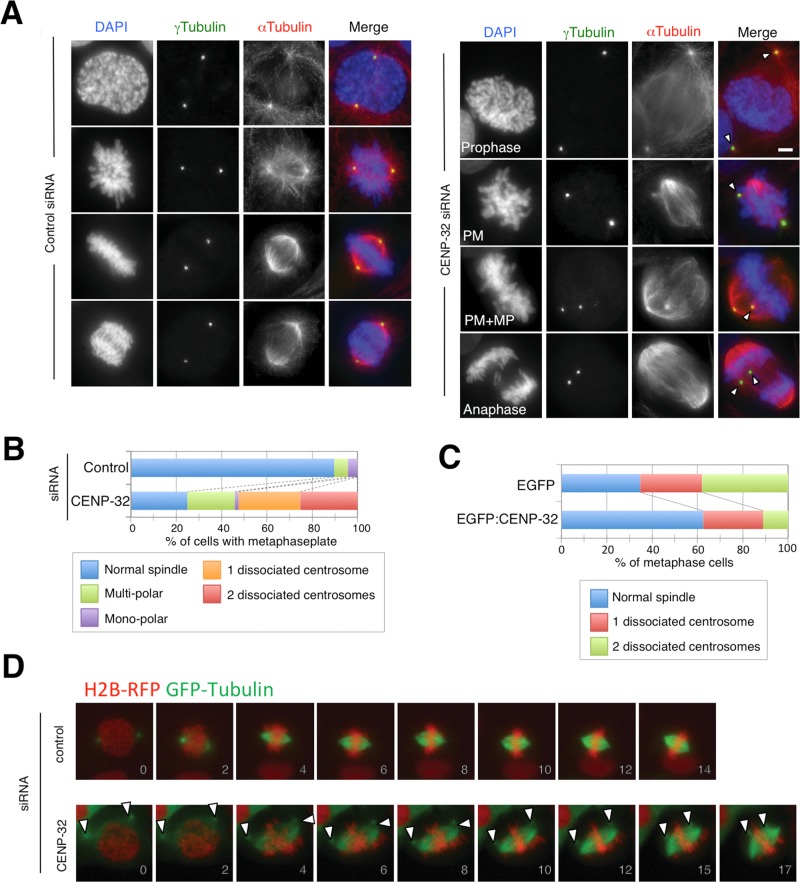
We previously reported that CENP-32 depletion resulted in the detachment of centrosomes from spindle poles during metaphase (Figure 1B; Ohta et al., 2010). To investigate the timing of this centrosomal dissociation, we used immunofluorescence staining against γ-tubulin and α-tubulin to visualize centrosomes and spindle microtubules, respectively (Figure 1A). Compared to cells treated with control small interfering RNA (siRNA), centrosomes of CENP-32 siRNA cells exhibited premature movement away from the nuclear surface before nuclear envelope breakdown (Figure 1A; prophase). Of interest, subsequent bipolar spindle formation was not abolished upon CENP-32 depletion, even though centrosomal detachment persisted after anaphase onset (Figure 1A; white arrowheads). Overexpression of enhanced green fluorescent protein (EGFP):CENP-32 reduced the percentage of centrosomal dissociation after CENP-32 siRNA treatment (Figure 1C). This rescue suggests that the centrosomal dissociation was caused by CENP-32 depletion.
FIGURE 1:
(A) Mitotic centrosome position in control and CENP-32 siRNA–transfected cells at prophase, prometaphase (PM), metaphase (or prometaphase with metaphase plate: PM + MP), and anaphase. Cells were immunolabeled with γ-tubulin (green) and α-tubulin (red) antibodies. DAPI staining was used to detect DNA (blue). (B) Quantification of phenotypes in CENP-32–depleted cells with metaphase plate. (C) Quantification of phenotypes in CENP-32–depleted cells with overexpression of EGFP (top) or EGFP:CENP32 (bottom). (D) Time-lapse analysis of control and CENP-32 siRNA–transfected cells. The number shows time (minutes) from nuclear envelope breakdown. White arrowheads indicate centrosomes dissociated from the mitotic spindle in A and D.
Time-lapse microscopy was also used to analyze spindle formation in CENP-32–depleted U2OS cells overexpressing GFP:α-tubulin and histone H2B:monomeric red fluorescent protein fusion proteins (Figure 1D). In CENP-32–depleted cells, detached centrosomes could be distinguished from the remainder of the bipolar spindle as isolated spots of microtubule staining (Figure 1D; arrow heads). Centrosomes appeared to dissociate from the mitotic spindle immediately after bipolar spindle formation in CENP-32 siRNA cells (Figure 1D). Despite this detachment, CENP-32 depletion did not have a dramatic effect on spindle morphology or chromosome alignment during prometaphase (Figure 1D).
These results reveal that CENP-32 is required for centrosome tethering at spindle poles and suggest that the protein is required for centrosomes to act as dominant polarity determinants of spindle pole formation.
Dissociated centrosomes have an altered protein composition
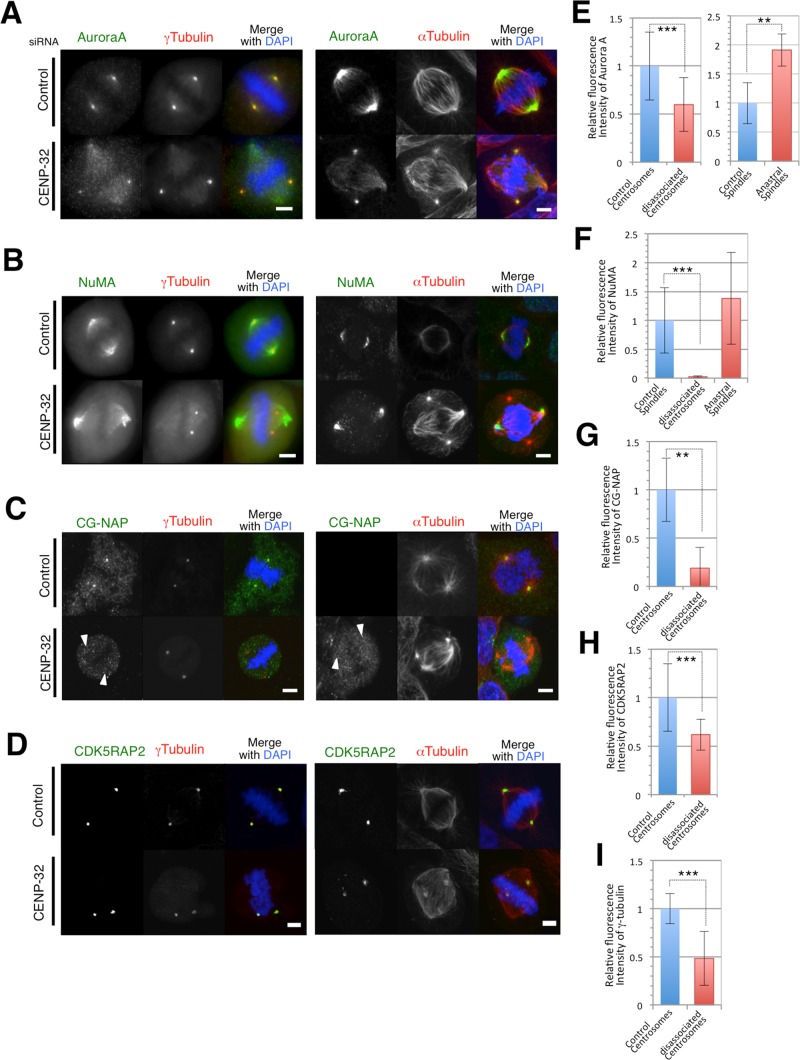
It was shown previously that dissociated centrosomes in CENP-32–depleted cells maintain γ-TuRC complex and pericentrin localization and that those proteins are absent from spindle poles after centrosome detachment (Ohta et al., 2010). Here we confirmed that γ-tubulin staining was present on detached centrosomes in CENP-32–depleted cells (Figure 1), albeit at decreased levels after their disassociation from the spindle poles (quantification in Figure 2I). In both control and CENP-32–depleted cells, weak γ-tubulin signals were also found on mitotic spindles. Double immunofluorescence labeling against the γ-TuRC complex and various centrosomal proteins was then used to assess the composition of the detached centrosomes in the depleted cells.
FIGURE 2:
Pericentrosomal protein localization (A, Aurora A; B, NuMA; C, CG-NAP; and D, CDK5RAP2) in control and CENP-32–depleted cells with DAPI and α-tubulin or γ-tubulin. (E–I) Quantification of immunofluorescence intensities of control and CENP-32 siRNA–transfected cells . **p < 0.05, ***p < 0.01.
Aurora A is required for centrosome maturation and bipolar spindle formation (Hannak et al., 2001). Compared to control cells, CENP-32 siRNA cells exhibited a weaker (but still positive) staining of Aurora A on the detached centrosomes and a stronger diffuse signal on spindle microtubules (Figure 2, A and E). In contrast, NuMA did not accumulate on the dissociated centrosomes but did continue to localize at the poles of the bipolar spindles (Figure 2, B and F). CG-NAP signals were also substantially decreased on detached centrosomes in CENP-32–depleted cells (Figure 2, C and G). It was previously reported that lack of CDK5RAP2 is able to cause centrosomal release from the spindle (Barr et al., 2010). However, CDK5RAP2 could still be detected on the detached centrosomes, albeit at decreased levels, in CENP-32–depleted cells (Figure 2, D and H).
CG-NAP knockdown does not affect centrosome dissociation from mitotic spindles
We next tested the hypothesis that CG-NAP mislocalization caused by CENP-32 depletion might serve as a trigger for centrosome dissociation from mitotic spindles. U2OS cells were depleted of CG-NAP by two different siRNA transient transfections (Supplemental Figure S1B). Immunofluorescence labeling against α-tubulin and pericentrin revealed that CG-NAP loss could occasionally result in a centrosome-dissociation phenotype in mitotic cells (Supplemental Figure S1C). However, this phenotype was observed at a much lower frequency than that found in CENP-32–depleted cells (Supplemental Figure S1, D and E; ∼6% in CG-NAP–depleted cells vs. ∼33% in CENP-32–depleted cells). CG-NAP depletion also resulted in premature centrosome movement away from the nuclear surface in ∼50% of prophase cells, similar to that observed in cells depleted of CENP-32 (unpublished data).
Thus, although loss of CG-NAP can explain certain aspects of the CENP-32–depletion phenotype, it cannot fully explain the dissociation of centrosomes from the spindle or their loss of polarity dominance.
Microtubule regrowth assay with CENP-32–depleted cells
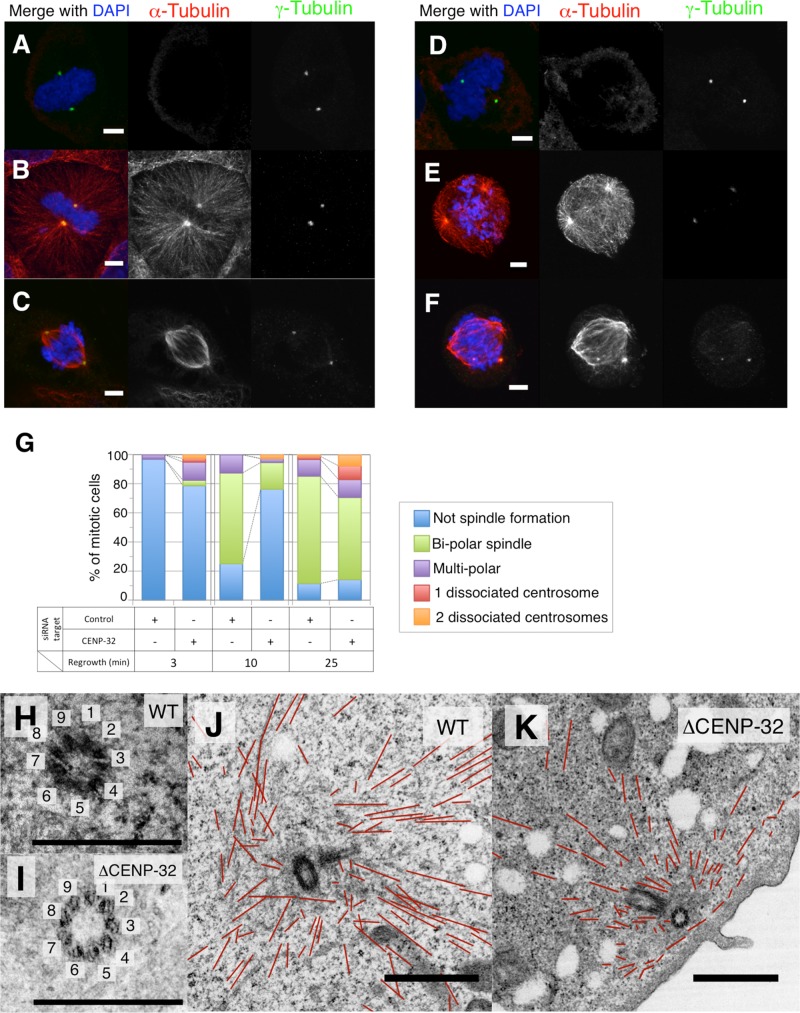
Next we examined the spindle formation ability of dissociated centrosomes, using a microtubule regrowth assay. Microtubules in control and CENP-32–depleted cells were disrupted by cold treatment for 30 min (Figure 3, A and D). In the control, microtubule initiation from centrosomes was observed by 3 min after the transfer to prewarmed medium (Figure 3B). After 25 min, 74% of cells showed bipolar spindles (Figure 3, C and G). In CENP-32–depleted cells, we observed significantly lower levels of microtubule polymerization from centrosomes by 3 min after the transfer to prewarmed medium (Figure 3E). Furthermore, 17% of cells showed anastral spindle after 25 min (Figure 3, F and G). These results suggest that the anastral spindle poles remain capable of nucleating microtubules after centrosome detachment, whereas nucleation by the detached centrosomes is significantly impaired.
FIGURE 3:
Immunostaining of α-tubulin (red) and γ-tubulin (green) after microtubule regrowth for 0 (A, D), 3 (B, E), and (C, F) 25 min. (G) Quantification of reformation of mitotic spindles in this assay. Observation of centrosomes under a transmission electron microscope of centrioles in control (H) and CENP-32 siRNA (I) cells. The number indicates the number of the triplet microtubules. Scale bar, 500 nm. (J) Microtubules around centrosomes in control and (K) CENP-32 siRNA cells. Scale bar, 1 μm. Red lines indicate observed microtubules.
The ultrastructure of dissociated centrosomes
Transmission electron microscopy was used to visualize the ultrastructure of dissociated centrosomes in CENP-32–depleted cells. Centrosomes in control and CENP-32–depleted cells contained centrioles with nine triplet microtubules assembled in a normal cartwheel structure (Figure 3, H and I). No major differences were observed in the structure of the centrioles or PCM surrounding dissociated centrosomes in comparison to normal, spindle pole–associated centrosomes (Figure 3J). However, we did observe an apparent decrease in the density of microtubules surrounding the dissociated centrosomes in CENP-32–depleted cells (Figure 3K). Furthermore, most microtubules associated with the detached centrosomes appeared to be shorter than those at control centrosomes. This observation is consistent with the impaired astral microtubule assembly observed in CENP-32–depleted cells in the microtubule regrowth assay.
Microtubule instability at spindle poles without centrosomes
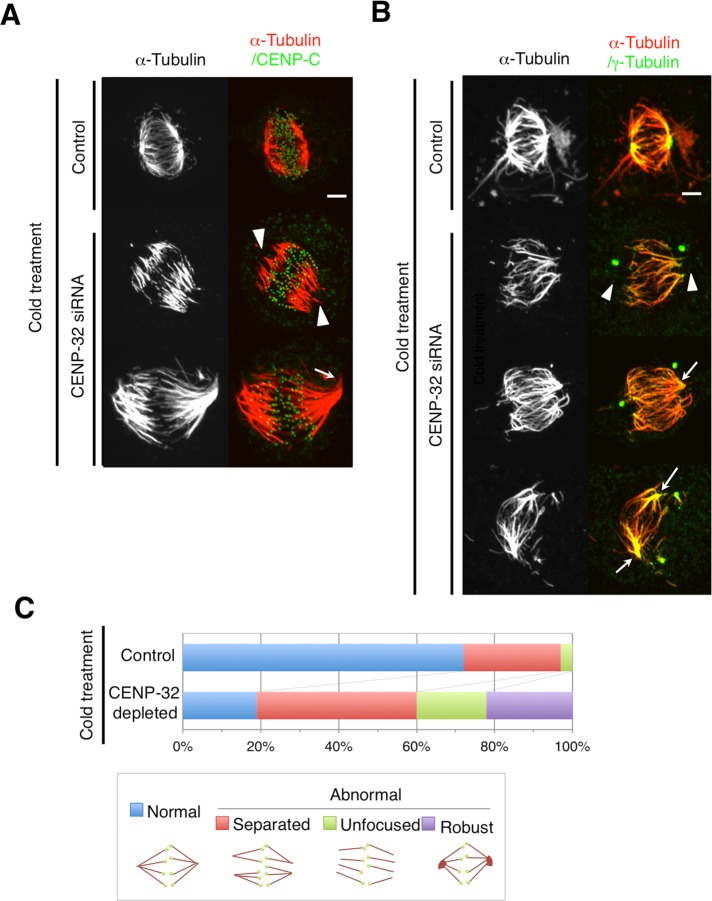
We next examined the stability of the spindle microtubules in cells with dissociated centrosomes induced by CENP-32 depletion. We did this by using a cold-stability assay. Under the conditions used, only stable spindle microtubules associated with kinetochores (K-fibers) withstand the cold treatment at 4°C for 10 min. After the cold treatment, cells were immunostained for the kinetochore marker, CENP-C, and α-tubulin to detect spindle morphology.
CENP-32 siRNA cells exhibited two distinct phenotypes after cold treatment. The first, observed in 18% of CENP-32–depleted cells, consisted of fragmented acentrosomal spindle poles (Figure 4, A and B, arrowheads, and C, unfocused). The second phenotype, observed in 22% of CENP-32–depleted cells after cold treatment, consisted of intact focused anastral spindle poles. These spindles contained robust microtubule bundles (Figure 4, A and B, white arrows, and C, robust).
FIGURE 4:
Depolymerization by cold treatment induces microtubule dissolution at spindle poles in CENP-32–depleted cells. (A) Cold-treated control (top) and CENP-32-depleted (bottom) cells are shown with α-tubulin (red) and CENP-C (green). (B) Cold-treated control (top) and CENP-32–depleted (bottom) cells are shown with α-tubulin (red) and γ-tubulin (green). Arrowheads indicate dissolution at spindle poles and robust microtubule bundles at spindle poles. (C) Quantification of four cell phenotypes observed in cold treatment experiments.
This experiment suggested that kinetochore–microtubule interactions were apparently stable after cold treatment in both control and CENP-32–depleted cells (Figure 4A). However, we were unable to observe any microtubule signal associated with the dissociated centrosomes in CENP-32–depleted cells, whereas a faint signal was seen in the cells exposed to control RNA interference (RNAi; Figure 4, A and B). Therefore the microtubules nucleated by the detached spindle poles apparently do not interact with kinetochores and also apparently interact more weakly with the detached centrosomes.
Taken together, the experiments in this and the preceding section reveal that CENP-32 is required for either the nucleation or stability of astral microtubules.
Augmin knockdown partially suppresses centrosome detachment in CENP-32–depleted cells
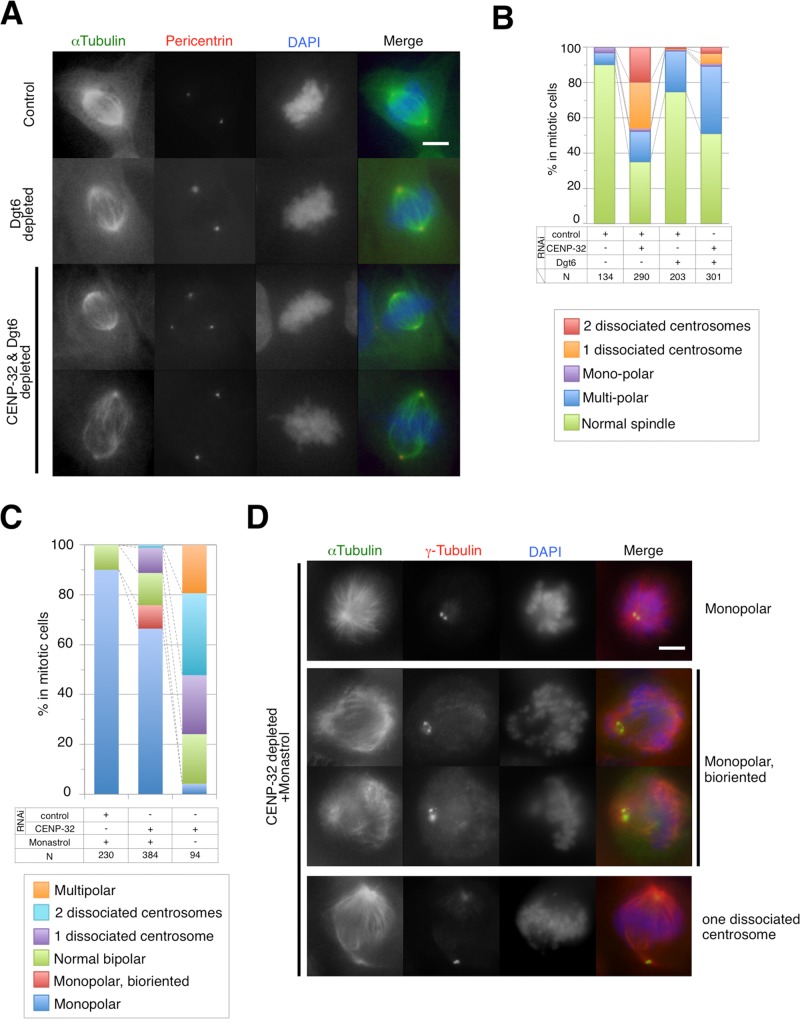
The augmin complex is required for the recruitment of the γ-TuRC to microtubules and promotes centrosome-independent microtubule formation (Uehara et al., 2009). To investigate the possibility that augmin is required for anastral spindle formation in CENP-32-depleted cells, we codepleted the augmin subunit Dgt6 using siRNA. As expected, spindles contained a reduced microtubule density after Dgt6 RNAi depletion (Figure 5A). Double staining for α-tubulin and pericentrin revealed that in single-target siRNA experiments, 33% of mitotic cells had more than one dissociated centrosome in CENP-32–depleted cells, whereas only 2% of mitotic cells exhibited this phenotype when Dgt6 levels were reduced (Figure 5B). We found that 5.6% of mitotic cells contained more than one dissociated centrosome in CENP-32 and Dgt6 double-depleted cells (Figure 5B). This result indicates that siRNA targeting of the augmin complex partly suppressed the centrosome-detachment phenotype in CENP-32–depleted cells. CENP-32 and Dgt6 double-depleted cells also exhibited a multipolar phenotype and prometaphase accumulation (Figure 5B; 37% of cells).
FIGURE 5:
Augmin depletion dissolves the centrosome-dissociation phenotype. (A) Dgt6 or Dgt6- and CENP-32–depleted cells show reduced mitotic spindle microtubule density. The most frequently observed spindles in Dgt6- and CENP-32–depleted cells are indicated with α-tubulin (green), pericentrin (red), and DAPI (blue). (B) Quantification of phenotypes in CENP-32– and Dgt6-depleted cells (bottom table). (C) Quantification of phenotypes in monastrol-treated CENP-32–depleted cells (bottom table). (D) The most frequently observed spindles in monastrol-treated CENP-32–depleted cells are indicated with a-Tubulin, g-Tubulin, and DAPI.
CENP-32 depletion causes a decrease in monopolar spindle formation accompanied by centrosome detachment
Bipolar spindles form as a result as balanced interactions between motor proteins, and imbalances in those forces can cause the spindles to collapse (Saunders and Hoyt, 1992; Saunders et al., 1997; Straight et al., 1998; Nazarova et al., 2013). A widely accepted approach used to look at the effect of disrupting this balance of forces involves the use of monastrol, an inhibitor of the bipolar kinesin-5 Eg5, during spindle assembly (Mayer et al., 1999). This inhibition results in efficient collapse of the spindle.
We found that although monopolar spindle formation still occurs with high efficiency in monastrol-treated, CENP-32–depleted cells, the process is less efficient than it is in control depleted cells (Figure 5C; 90% in control and 66% in CENP-32 depleted). The background of normal bipolar spindles observed after monastrol treatment was unchanged by CENP-32 depletion (Figure 5C; 10% in control and 13% in CENP-32 depleted).
Of interest, the decrease in monopolar spindles observed in the CENP-32–depleted cells was balanced primarily by an increase in bipolar, monoastral spindles (Figure 5C; not observed in controls, but 11% in CENP-32 depleted). CENP-32 depletion and monastrol treatment also resulted in the production of 9% of cells with a remarkable phenotype in which both centrosomes were clustered as a single pole but the majority of chromosomes appeared to be bioriented on a metaphase plate.
We hypothesize that in the 11% of cells with bipolar, monoastral spindles, the forces that normally cause spindle collapse resulted in detachment of one centrosome from a spindle pole, and this blocked the process of spindle collapse.
NuMA and CENP-32 double-depleted cells are not able to maintain anastral bipolar spindles
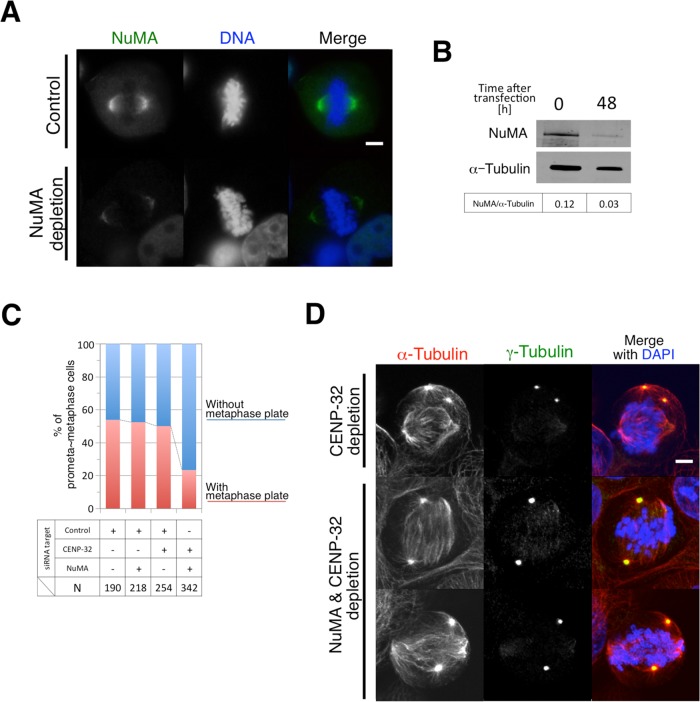
NuMA localized at the poles of the bipolar spindles in CENP-32–depleted cells (Figure 1B). We predicted that NuMA might tether the anastral spindle poles in CENP-32–depleted cells as it does in Xenopus egg extracts (Merdes et al., 1996) and that if NuMA is depleted in addition to CENP-32, the integrity of the anastral spindles might not be maintained.
Single siRNA treatment targeting NuMA depleted 75% of endogenous NuMA (Figure 6, A and B). Double depletion of NuMA and CENP-32 did not suppress the centrosome detachment seen in metaphase cells after CENP-32 depletion (Supplemental Figure S2A; CENP-32 depletion, 48%; NuMA plus CENP-32 double depletion, 54%). To investigate whether double-depleted cells are able to construct functional bipolar spindles and align chromosomes at a metaphase plate, we determined the frequency of metaphase cells in cultures after double depletion of NuMA and CENP-32 or single depletion of CENP-32 (Figure 6D). This analysis showed that double depletion of NuMA and CENP-32 caused a marked decrease in the frequency of cells with a metaphase plate after CENP-32 depletion (Figure 6C and Supplemental Figure S2B; CENP-32 depletion, 46.9%; NuMA plus CENP-32 double depletion, 21.4%).
FIGURE 6:
NuMA depletion does not affect centrosome dissociation. (A) NuMA siRNA transfection leads to NuMA mislocalization at centrosomes. NuMA (green), DAPI (blue), and merged. (B) Immunoblots of CENP-32 siRNA–transfected cell extracts after 0 and 48 h. (C) Quantification of the mitotic cells with/without metaphase plate. (D) The most frequently observed spindles in Dgt6- and CENP-32–depleted cells are indicated with α-tubulin, γ-tubulin, and DAPI.
CENP-32 is also important for spindle organization in chicken mitotic cells
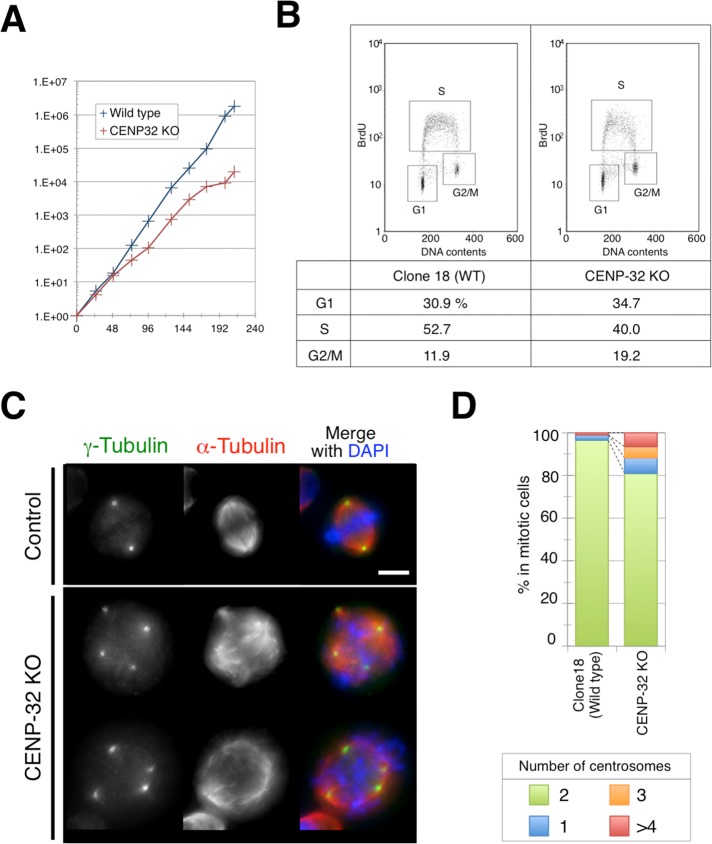
We generated a CENP-32 deletion mutant in chicken DT40 cells (Supplemental Figure S3). Of interest, this mutation was not lethal to chicken cells but did induce a mild growth defect. Compared to wild-type cells (doubling time of 10.7 h), loss of CENP-32 caused the doubling time to increase to 14.5 h (Figure 7A). CENP-32 depletion also caused a small increase in the percentage of cells in the G2/M phase (19.2% in the CENP-32 knockout compared with 11.9% for wild-type cells) as measured by fluorescence-activated cell sorting (FACS) analysis (Figure 7B). However, CENP-32 depletion did not cause a significant increase in the percentage of cells phosphorylated at serine 10 in histone H3 (1.9% in the CENP-32 knockout and 1.7% in wild-type cells; Supplemental Figure S4) as measured by FACS analysis. The increase in G2/M cells may have been partly explained by the fact that CENP-32–knockout cells exhibited an increase in number of cells with multipolar spindles (from 3.5% in controls to 18.8%: Figure 7, C and D).
FIGURE 7:
CENP-32 knockout in chicken DT40 cells shows multipolar phenotype.(A) Growth curves of wild-type (blue) and CENP-32–knockout (red) cells. (B) BrdU and propidium iodide (PI) plots are analyzed by FACS. The windows for separated G1, S, and G2/M phases are indicated. Quantification is shown at the bottom. (C) Typical CENP-32 phenotype is a multipolar spindle merged with DAPI (blue), γ-tubulin (green), and α-tubulin (red). (D) Quantification of observed CENP-32 phenotypes. In three independent experiments, we quantified a total of 306 mitotic wild-type (clone-18 cells) and 387 mitotic CENP-32–knockout cells.
DISCUSSION
Time-lapse microscopy analysis revealed that CENP-32 depletion resulted in centrosome dissociation from mitotic spindles during early prometaphase. After centrosome dissociation, the mitotic spindles remained active and promoted chromosome alignment to a metaphase plate. Moreover, these mitotic spindles without centrosomes (“anastral” spindles) were able to advance to anaphase. Remarkably, the detached centrosomes appeared to have lost the ability to act as dominant determinants of spindle polarity. Although they could still nucleate microtubules, the efficiency of microtubule nucleation and/or the stability of those associated microtubules were significantly less than normal. As a result, the detached centrosomes appeared no longer able to integrate structurally into the body of the spindle—indeed, they often migrated across the surface of the spindle, moving toward the metaphase plate without disturbing the underlying spindle structure.
CENP-32 knockdown results in loss of CG-NAP from centrosomes
CG-NAP failed to accumulate on the dissociated centrosomes after CENP-32 depletion, even though the γ-TuRC and pericentrin were present. Furthermore, centrosomes in both CENP-32–depleted and CG-NAP–depleted cells exhibited premature separation from the nuclear envelope during prophase. However, depletion of CG-NAP alone did not result in centrosome detachment from the body of the spindle. Thus CG-NAP appears to be downstream of CENP-32 in this process.
CG-NAP is important for centrosome maturation (Takahashi et al., 1999, 2002). We speculate that CENP-32 may promote centrosome maturation by controlling CG-NAP localization to centrosomes in early mitosis. Our data suggest that not only centrosome targeting of CG-NAP but also other structural changes resulting from CENP-32 depletion must contribute to centrosome dissociation from mitotic spindle poles.
Mitotic spindle poles without centrosomes are tethered by NuMA
The fact that chromosomes were still able to align at a metaphase plate after centrosome detachment in CENP-32–depleted cells suggests that the basic functionality of the anastral mitotic spindle is maintained even in the absence of centrosomes. Of importance, both a microtubule regrowth assay and electron microscope observations suggested that microtubule elongation from detached centrosomes is impaired in cells with anastral spindles. In contrast, a cold-stable microtubule depolymerization assay showed that kinetochore–microtubule interactions remained robust in CENP-32–depleted cells. This therefore raises the question of how the anastral spindles in CENP-32–knockdown cells are maintained.
NuMA was previously shown to interact with dynein to promote spindle pole formation in Xenopus egg extracts by cooperating with cytoplasmic dynein to cross-link parallel microtubules (Merdes et al., 1996, 2000). Furthermore, after CENP-32 depletion, NuMA remained at the poles of the anastral spindles and did not associate with the dissociated centrosomes. Bipolar spindle morphology was severely compromised after double depletion of CENP-32 and NuMA. Thus NuMA is one of the factors that maintain the anastral spindles in CENP-32–depleted cells.
Factors that form and maintain anastral mitotic spindles
Centrosomal dissociation from the mitotic spindle was previously reported in CDK5RAP2-knockdown cells (Barr et al., 2010). In fact, our indirect immunofluorescence microscopy analysis showed CDK5RAP2 remained on disassociated centrosomes in CENP-32– depleted cells, albeit at lower levels (Figure 1D). This result suggests that CENP-32 might anchor centrosomes at mitotic spindle poles via a mechanism different from CDK5RAP2, although it is possible that the decrease in centrosome-associated CDKRAP2 levels after CENP-32 depletion is sufficient to promote centrosome detachment. Augmin knockdown was able to suppress this centrosomal-dissociation phenotype (Moutinho-Pereira et al., 2013). Moreover, double knockdown of CENP-32 and Dgt6 (augmin) partly suppressed the centrosome detachment phenotype seen after CENP-32 depletion. Augmin recruits γ-TuRC to microtubule walls and contributes to centrosome-independent microtubule generation (Goshima et al., 2008; Kamasaki et al., 2013). It was recently reported that augmin is involved in centrosome-independent mitotic spindle assembly in Drosophila S3 cells and plants (Ho et al., 2011; Hotta et al., 2012; Hashimoto, 2013; Moutinho-Pereira et al., 2013).
One role of augmin is apparently to make the central portion of the spindle more robust. This could create a functional junction between the aster and the central region of the spindle. Spindle forces acting after loss of CENP-32 might cause a weakened junction to break, thereby releasing the centrosome and leaving behind an anastral spindle. In the absence of augmin, the less robust central spindle might be more flexible, so that the centrosome does not detach when the same forces act.
Alternatively, γ-TuRC levels might potentially increase at centrosomes after augmin depletion. This could potentially increase their microtubule nucleation ability, thereby integrating them more robustly into the body of the spindle. However, note that under the conditions used here, Dgt6 knockdown did not cause an increase in γ-TuRC levels at spindle poles as measured by indirect immunofluorescence.
It is also possible that the presence of augmin is somehow required in ordered for the forces that occur after depletion of CENP-32 to cause spindle detachment. The nature of those forces is unknown, but it has been shown that depletion of dynein subunits in Drosophila cells can result in centrosome detachment from spindles (Morales-Mulia and Scholey, 2005). Of note, those spindle poles became unfocused, and the spindles were significantly longer than wild type, phenotypes that are occasionally seen in the case of CENP-32 depletion. An alternative explanation is that some factor downstream of the large scaffolding protein CG-NAP is involved in linking centrosomes to the central region of the spindle.
That forces within the spindle cause the centrosome detachment is also suggested by the observation that CENP-32 depletion results in a small but significant decrease in the formation of monopolar spindles in monastrol-treated cells. This decrease is balanced by an increase in the number of bipolar, monoastral spindles, suggesting that in those cells, the spindle forces normally counteracted by Eg5 cause the detachment of a centrosome from one pole instead of pulling the two poles together.
CENP-32 and centrosomal dominance in spindle assembly
Even though spindles can form perfectly well in the absence of centrosomes (e.g.. in eggs and higher plants, as well as in vertebrate cultured cells after experimental manipulations), it is generally believed that where they are present, centrosomes act as dominant determinants of spindle pole formation. Remarkably, the microtubules associated with CENP-32–detached centrosomes do not make stable attachments to kinetochores and appear not to insinuate themselves into the body of the spindle. These results suggest that microtubules associated with detached centrosomes in CENP-32–depleted cells behave like astral microtubules rather than like components of the central body of the spindle. This is despite the fact that the detached centrosomes retain both Aurora A and γ-tubulin. Thus CENP-32 is apparently required for centrosomes to behave like spindle poles capable of nucleating both spindle and astral microtubules.
Future experiments with CENP-32–depleted cells may allow identification of the determinants that enable centrosomes to act as dominant determinants of spindle pole formation.
MATERIALS AND METHODS
Cell culture
U2OS cells in exponential growth were seeded onto coverslips and grown overnight at 37°C in RPMI/10% fetal bovine serum (FBS) at 5% CO2. DT40 cells with the CENP-32 mutant were maintained in RPMI 1640 medium supplemented with 10% (vol/vol) FBS, 1% chicken serum (Life Technologies, Grand Island, NY), 100 U/ml penicillin, 100 μg/ml streptomycin (Wako, Osaka, Japan) at 39°C, and 5% CO2 in a humid incubator.
Drug treatment
Monastrol (Sigma-Aldrich, St. Louis, MO) was dissolved in dimethyl sulfoxide (DMSO) and used at a final concentration of 68 M for 12 h. DMSO was added to mock-treated controls.
Transfection and indirect immunofluorescence microscopy
A 100-pmol amount of siRNA (control, AACGUACGCGGAAUACUUCGAdTdT; CENP-32, GCAGGACCCUCGCACCAAAdTdT, Ohta et al., 2010; CG-NAP-si1, GCUUCUAUUUAGUCACGAAdTdT, Ozaki et al., 2012); CG-NAP-si2, GCAUGGAUGCUUCUAGACAdTdT; Dgt6, CAGUUAAGCAGGUACGAAAdTdT, Uehara and Goshima, 2010); NuMA, CUAGCUGAGCUCCAUGCCAdTdT, Haren et al., 2009) was administered to U2OS cells at 30–40% confluence by transfection with Oligofectamine or Lipofectamine RNAi MAX (Life Technologies) in complete medium without antibiotics. To rescue siRNA for CENP-32, pDEST131NEGFP-CENP32 or pDEST131NEGFP-CTR was transfected to U2OS cells at 30–40% confluence using Lipofectamine LTX (Life Technologies) before 12 h from siRNA treatment. Cells maintained in this medium for 48–72 h were fixed for 5 min with 4% (vol/vol) paraformaldehyde (Electron Microscopy Services) in phosphate-buffered saline (PBS) for 2 min with cold methanol or for 10 min with methanol/acetone at −20°C. After permeabilization with 0.15% (vol/vol) Triton X-100 in PBS, coverslips were blocked with 1% (vol/vol) BSA in PBS. Cells were probed with antibodies against CENP-C (1:600, rabbit 554), α-tubulin (1:2000, B512; Sigma-Aldrich), γ-tubulin (1:1000, AK15, Sigma-Aldrich; 1:1000, GTU-88, Sigma-Aldrich), pericentrin (1:1000, ab4448; Abcam, Cambridge, UK), CG-NAP (1:500; Takahashi et al., 1999), Aurora A (1:1000, ab13824, Abcam; 1:100, #4718, Cell Signaling Technology, Danvers, MA), NuMA (1:100, #3888; Cell Signaling Technology), or CDK5RAP2 (1:1000, #06-1398; Millipore, Billerica, MA). Cells were washed three times with PBS for 5 min, Alexa-conjugated secondary antibodies were applied at 1:600, and the DNA was counterstained with 4′,6-diamidino-2-phenylindole (DAPI) at 0.1 μg/ml.
Microtubule regrowth assay
Microtubule regrowth was induced by replacing the cold DMEM with prewarmed medium at 37°C. At the indicated time points after medium replacement, cells were fixed by cold methanol and immunostained with indicated antibody and Alexa-conjugated secondary antibody.
Cold treatment assay
Cells were treated in ice-cold DMEM for 10 min after ice-cold PHME (50 mM PIPES, 25 mM HEPES, 10 mM EGTA, 4 mM MgSO4, and 0.5% Triton X-100) buffer treatment for 2 min. Cells were fixed with cold Me-OH for 2 min and washed twice with PBS for 15 min. Immunofluorescence was done as described above.
Immunoblotting
Primary antibodies used for immunoblotting were rabbit anti-NuMA at 1:10000, rabbit anti–CG-NAP at 1:1000, and mouse anti–a-tubulin at 1:2000. The secondary antibodies were IRDye 800CW donkey anti–a-rabbit immunoglobulin IgG at 1:10,000 and anti-mouse IgG at 1:10,000 (926-32213, 926-32210; LI-COR Biosciences, Lincoln, NE).
Electron microscopy
The culture cells were fixed with 1% glutaraldehyde in PBS, pH 7.3, for 1 h at 4°C. They were then postfixed with 1.5% osmium tetroxide (or osmic acid) in 0.1 M phosphate buffer, pH 7.3. for 1 h at 4°C and dehydrated in a graded series of ethanol. after dehydration, they were embedded in Epon 812 (TAAB Laboratories Equipment, Berkshire, United Kingdom). They were observed with a JEM-1400Plus electron microscope (JEOL, Tokyo, Japan).
FACS
FACS analysis was carried out on DT40 cells maintained in media containing bromodeoxyuridine (BrdU) for 30 min before harvest. Cells were washed once in PBS with 1% BSA and fixed in 70% ethanol. The fixed cells were washed again once with PBS and probed with anti-BrdU fluorescein isothiocyanate (1:100, 11-5071; eBioscience, San Diego, CA). The cells were washed with PBS and treated with propidium iodide and RNase A for 30 min. The samples were analyzed through flow cytometry using BD FACSCalibur.
Supplementary Material
Acknowledgments
We thank Mikiko Takahashi (Teikyo Heisei University, Tokyo, Japan) for CG-NAP antibody. We thank Shuji Sakamoto, Mikiro Takaishi, and Takuma Higuchi (Kochi University, Kochi, Japan) for critical reading of the manuscript. This work was supported by Japan Society for the Promotion of Science KAKENHI Grant 25870487 to S.O. and Wellcome Trust Principal Research Fellowship Grant 073915 to W.C.E. The Wellcome Trust Centre for Cell Biology is supported by Core Grants 077707 and 092076.
Abbreviations used:
- CENP-32
centromere protein 32
- CG-NAP
centrosome and Golgi–localized, PKN-associated protein
- Dgt2-9
dim γ-tubulin 2-9
- PCM
pericentriolar material
- γ-TuRC
γ-tubulin ring complex.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E14-09-1366) on February 5, 2015.
REFERENCES
- Barr AR, Kilmartin JV, Gergely F. CDK5RAP2 functions in centrosome to spindle pole attachment and DNA damage response. J Cell Biol. 2010;189:23–39. doi: 10.1083/jcb.200912163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrera JA, Kao L-R, Hammer RE, Seemann J, Fuchs JL, Megraw TL. CDK5RAP2 regulates centriole engagement and cohesion in mice. Dev Cell. 2010;18:913–926. doi: 10.1016/j.devcel.2010.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bird SL, Heald R, Weis K. RanGTP and CLASP1 cooperate to position the mitotic spindle. Mol Biol Cell. 2013;24:2506–2514. doi: 10.1091/mbc.E13-03-0150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byers B, Goetsch L. Duplication of spindle plaques and integration of the yeast cell cycle. Cold Spring Harb Symp Quant Biol. 1974;38:123–131. doi: 10.1101/sqb.1974.038.01.016. [DOI] [PubMed] [Google Scholar]
- Casenghi M, Meraldi P, Weinhart U, Duncan PI, Körner R, Nigg EA. Polo-like kinase 1 regulates Nlp, a centrosome protein involved in microtubule nucleation. Dev Cell. 2003;5:113–125. doi: 10.1016/s1534-5807(03)00193-x. [DOI] [PubMed] [Google Scholar]
- Dictenberg JB, Zimmerman W, Sparks CA, Young A, Vidair C, Zheng Y, Carrington W, Fay FS, Doxsey SJ. Pericentrin and gamma-tubulin form a protein complex and are organized into a novel lattice at the centrosome. J Cell Biol. 1998;141:163–174. doi: 10.1083/jcb.141.1.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doxsey S, McCollum D, Theurkauf W. Centrosomes in cellular regulation. Annu Rev Cell Dev Biol. 2005;21:411–434. doi: 10.1146/annurev.cellbio.21.122303.120418. [DOI] [PubMed] [Google Scholar]
- Dunsch AK, Hammond D, Lloyd J, Schermelleh L, Gruneberg U, Barr FA. Dynein light chain 1 and a spindle-associated adaptor promote dynein asymmetry and spindle orientation. J Cell Biol. 2012;198:1039–1054. doi: 10.1083/jcb.201202112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espiritu EB, Krueger LE, Ye A, Rose LS. CLASPs function redundantly to regulate astral microtubules in the C. elegans embryo. Dev Biol. 2012;368:242–254. doi: 10.1016/j.ydbio.2012.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fong K-W, Choi Y-K, Rattner JB, Qi RZ. CDK5RAP2 is a pericentriolar protein that functions in centrosomal attachment of the gamma-tubulin ring complex. Mol Biol Cell. 2008;19:115–125. doi: 10.1091/mbc.E07-04-0371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaglio T, Saredi A, Compton DA. NuMA is required for the organization of microtubules into aster-like mitotic arrays. J Cell Biol. 1995;131:693–708. doi: 10.1083/jcb.131.3.693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goshima G, Mayer M, Zhang N, Stuurman N, Vale RD. Augmin: a protein complex required for centrosome-independent microtubule generation within the spindle. J Cell Biol. 2008;181:421–429. doi: 10.1083/jcb.200711053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hachet V, Canard C, Gönczy P. Centrosomes promote timely mitotic entry in C. elegans embryos. Dev Cell. 2007;12:531–541. doi: 10.1016/j.devcel.2007.02.015. [DOI] [PubMed] [Google Scholar]
- Hannak E, Kirkham M, Hyman AA, Oegema K. Aurora-A kinase is required for centrosome maturation in Caenorhabditis elegans. J Cell Biol. 2001;155:1109–1116. doi: 10.1083/jcb.200108051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haren L, Gnadt N, Wright M, Merdes A. NuMA is required for proper spindle assembly and chromosome alignment in prometaphase. BMC Res Notes. 2009;2:64. doi: 10.1186/1756-0500-2-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto T. A ring for all: γ-tubulin-containing nucleation complexes in acentrosomal plant microtubule arrays. Curr Opin Plant Biol. 2013;16:698–703. doi: 10.1016/j.pbi.2013.09.002. [DOI] [PubMed] [Google Scholar]
- Heald R, Tournebize R, Blank T, Sandaltzopoulos R, Becker P, Hyman A, Karsenti E. Self-organization of microtubules into bipolar spindles around artificial chromosomes in Xenopus egg extracts. Nature. 1996;382:420–425. doi: 10.1038/382420a0. [DOI] [PubMed] [Google Scholar]
- Ho C-MK, Hotta T, Kong Z, Zeng CJT, Sun J, Lee Y-RJ, Liu B. Augmin plays a critical role in organizing the spindle and phragmoplast microtubule arrays in Arabidopsis. Plant Cell. 2011;23:2606–2618. doi: 10.1105/tpc.111.086892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hotta T, Kong Z, Ho C-MK, Zeng CJT, Horio T, Fong S, Vuong T, Lee Y-RJ, Liu B. Characterization of the Arabidopsis augmin complex uncovers its critical function in the assembly of the acentrosomal spindle and phragmoplast microtubule arrays. Plant Cell. 2012;24:1494–1509. doi: 10.1105/tpc.112.096610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hung LY, Tang CJ, Tang TK. Protein 4.1 R-135 interacts with a novel centrosomal protein. (CPAP) which is associated with the gamma-tubulin complex. Mol Cell Biol. 2000;20:7813–7825. doi: 10.1128/mcb.20.20.7813-7825.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackman M, Lindon C, Nigg EA, Pines J. Active cyclin B1-Cdk1 first appears on centrosomes in prophase. Nat Cell Biol. 2003;5:143–148. doi: 10.1038/ncb918. [DOI] [PubMed] [Google Scholar]
- Jia Y, Fong K-W, Choi Y-K, See S-S, Qi RZ. Dynamic recruitment of CDK5RAP2 to centrosomes requires its association with dynein. PLoS One. 2013;8:e68523. doi: 10.1371/journal.pone.0068523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalab P, Weis K, Heald R. Visualization of a Ran-GTP gradient in interphase and mitotic Xenopus egg extracts. Science. 2002;295:2452–2456. doi: 10.1126/science.1068798. [DOI] [PubMed] [Google Scholar]
- Kamasaki T, O'Toole E, Kita S, Osumi M, Usukura J, McIntosh JR, Goshima G. Augmin-dependent microtubule nucleation at microtubule walls in the spindle. J Cell Biol. 2013;202:25–33. doi: 10.1083/jcb.201304031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karsenti E, Newport J, Kirschner M. Respective roles of centrosomes and chromatin in the conversion of microtubule arrays from interphase to metaphase. J Cell Biol. 1984;99:47s–54s. doi: 10.1083/jcb.99.1.47s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khodjakov A, Cole RW, Oakley BR, Rieder CL. Centrosome-independent mitotic spindle formation in vertebrates. Curr Biol. 2000;10:59–67. doi: 10.1016/s0960-9822(99)00276-6. [DOI] [PubMed] [Google Scholar]
- Kimura M, Kotani S, Hattori T, Sumi N, Yoshioka T, Todokoro K, Okano Y. Cell cycle-dependent expression and spindle pole localization of a novel human protein kinase, Aik, related to Aurora of Drosophila and yeast Ipl1. J Biol Chem. 1997;272:13766–13771. doi: 10.1074/jbc.272.21.13766. [DOI] [PubMed] [Google Scholar]
- Kirschner M, Mitchison T. Beyond self-assembly: from microtubules to morphogenesis. Cell. 1986;45:329–342. doi: 10.1016/0092-8674(86)90318-1. [DOI] [PubMed] [Google Scholar]
- Kiyomitsu T, Cheeseman IM. Chromosome- and spindle-pole-derived signals generate an intrinsic code for spindle position and orientation. Nat Cell Biol. 2012;14:311–317. doi: 10.1038/ncb2440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiyomitsu T, Cheeseman IM. Cortical dynein and asymmetric membrane elongation coordinately position the spindle in anaphase. Cell. 2013;154:391–402. doi: 10.1016/j.cell.2013.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laan L, Pavin N, Husson J, Romet-Lemonne G, van Duijn M, López MP, Vale RD, Jülicher F, Reck-Peterson SL, Dogterom M. Cortical dynein controls microtubule dynamics to generate pulling forces that position microtubule asters. Cell. 2012;148:502–514. doi: 10.1016/j.cell.2012.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lüders J, Stearns T. Microtubule-organizing centres: a re-evaluation. Nat Rev Mol Cell Biol. 2007;8:161–167. doi: 10.1038/nrm2100. [DOI] [PubMed] [Google Scholar]
- Maiato H, Khodjakov A, Rieder CL. Drosophila CLASP is required for the incorporation of microtubule subunits into fluxing kinetochore fibres. Nat Cell Biol. 2005;7:42–47. doi: 10.1038/ncb1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masoud K, Herzog E, Chabouté M-E, Schmit A-C. Microtubule nucleation and establishment of the mitotic spindle in vascular plant cells. Plant J. 2013;75:245–257. doi: 10.1111/tpj.12179. [DOI] [PubMed] [Google Scholar]
- Mayer TU, Kapoor TM, Haggarty SJ, King RW, Schreiber SL, Mitchison TJ. Small molecule inhibitor of mitotic spindle bipolarity identified in a phenotype-based screen. Science. 1999;286:971–974. doi: 10.1126/science.286.5441.971. [DOI] [PubMed] [Google Scholar]
- Merdes A, Heald R, Samejima K, Earnshaw WC, Cleveland DW. Formation of spindle poles by dynein/dynactin-dependent transport of NuMA. J Cell Biol. 2000;149:851–862. doi: 10.1083/jcb.149.4.851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merdes A, Ramyar K, Vechio JD, Cleveland DW. A complex of NuMA and cytoplasmic dynein is essential for mitotic spindle assembly. Cell. 1996;87:447–458. doi: 10.1016/s0092-8674(00)81365-3. [DOI] [PubMed] [Google Scholar]
- Miller L, Dai Q, Vegesna A, Korimilli A, Ulerich R, Schiffner B, Brassuer J. A missing sphincteric component of the gastro-oesophageal junction in patients with GORD. Neurogastroenterol Motil. 2009;21:813–e852. doi: 10.1111/j.1365-2982.2009.01294.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mimori-Kiyosue Y, Grigoriev I, Lansbergen G, Sasaki H, Matsui C, Severin F, Galjart N, Grosveld F, Vorobjev I, Tsukita S, et al. CLASP1 and CLASP2 bind to EB1 and regulate microtubule plus-end dynamics at the cell cortex. J Cell Biol. 2005;168:141–153. doi: 10.1083/jcb.200405094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchison TJ, Kirschner MW. Properties of the kinetochore in vitro. II. Microtubule capture and ATP-dependent translocation. J Cell Biol. 1985;101:766–777. doi: 10.1083/jcb.101.3.766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morales-Mulia S, Scholey JM. Spindle pole organization in Drosophila S2 cells by dynein, abnormal spindle protein (Asp), KLP10A. Mol Biol Cell. 2005;16:3176–3186. doi: 10.1091/mbc.E04-12-1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moutinho-Pereira S, Stuurman N, Afonso O, Hornsveld M, Aguiar P, Goshima G, Vale RD, Maiato H. Genes involved in centrosome-independent mitotic spindle assembly in Drosophila S2 cells. Proc Natl Acad Sci USA. 2013;110:19808–19813. doi: 10.1073/pnas.1320013110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nazarova E, O'Toole E, Kaitna S, Francois P, Winey M, Vogel J. Distinct roles for antiparallel microtubule pairing and overlap during early spindle assembly. Mol Biol Cell. 2013;24:3238–3250. doi: 10.1091/mbc.E13-05-0232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Connell CB, Khodjakov AL. Cooperative mechanisms of mitotic spindle formation. J Cell Sci. 2007;120:1717–1722. doi: 10.1242/jcs.03442. [DOI] [PubMed] [Google Scholar]
- O'Connell CB, Wang YL. Mammalian spindle orientation and position respond to changes in cell shape in a dynein-dependent fashion. Mol Biol Cell. 2000;11:1765–1774. doi: 10.1091/mbc.11.5.1765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohta S, Bukowski-Wills J-C, Sanchez-Pulido L, Alves F de L, Wood L, Chen ZA, Platani M, Fischer L, Hudson DF, Ponting CP, et al. The protein composition of mitotic chromosomes determined using multiclassifier combinatorial proteomics. Cell. 2010;142:810–821. doi: 10.1016/j.cell.2010.07.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oshimori N, Li X, Ohsugi M, Yamamoto T. Cep72 regulates the localization of key centrosomal proteins and proper bipolar spindle formation. EMBO J. 2009;28:2066–2076. doi: 10.1038/emboj.2009.161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oshimori N, Ohsugi M, Yamamoto T. The Plk1 target Kizuna stabilizes mitotic centrosomes to ensure spindle bipolarity. Nat Cell Biol. 2006;8:1095–1101. doi: 10.1038/ncb1474. [DOI] [PubMed] [Google Scholar]
- Ozaki Y, Matsui H, Asou H, Nagamachi A, Aki D, Honda H, Yasunaga S, Takihara Y, Yamamoto T, Izumi S, et al. Poly-ADP ribosylation of Miki by tankyrase-1 promotes centrosome maturation. Mol Cell. 2012;47:694–706. doi: 10.1016/j.molcel.2012.06.033. [DOI] [PubMed] [Google Scholar]
- Patel K, Nogales E, Heald R. Multiple domains of human CLASP contribute to microtubule dynamics and organization in vitro and in Xenopus egg extracts. Cytoskeleton (Hoboken) 2012;69:155–165. doi: 10.1002/cm.21005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Portier N, Audhya A, Maddox PS, Green RA, Dammermann A, Desai A, Oegema K. A microtubule-independent role for centrosomes and aurora a in nuclear envelope breakdown. Dev Cell. 2007;12:515–529. doi: 10.1016/j.devcel.2007.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reis R, Feijão T, Gouveia S, Pereira AJ, Matos I, Sampaio P, Maiato H, Sunkel CE. Dynein and mast/orbit/CLASP have antagonistic roles in regulating kinetochore-microtubule plus-end dynamics. J Cell Sci. 2009;122:2543–2553. doi: 10.1242/jcs.044818. [DOI] [PubMed] [Google Scholar]
- Samora CP, Mogessie B, Conway L, Ross JL, Straube A, McAinsh AD. MAP4 and CLASP1 operate as a safety mechanism to maintain a stable spindle position in mitosis. Nat Cell Biol. 2011;13:1040–1050. doi: 10.1038/ncb2297. [DOI] [PubMed] [Google Scholar]
- Saunders WS, Hoyt MA. Kinesin-related proteins required for structural integrity of the mitotic spindle. Cell. 1992;70:451–458. doi: 10.1016/0092-8674(92)90169-d. [DOI] [PubMed] [Google Scholar]
- Saunders W, Lengyel V, Hoyt MA. Mitotic spindle function in Saccharomyces cerevisiae requires a balance between different types of kinesin-related motors. Mol Biol Cell. 1997;8:1025–1033. doi: 10.1091/mbc.8.6.1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawin KE, LeGuellec K, Philippe M, Mitchison TJ. Mitotic spindle organization by a plus-end-directed microtubule motor. Nature. 1992;359:540–543. doi: 10.1038/359540a0. [DOI] [PubMed] [Google Scholar]
- Sharp DJ, Brown HM, Kwon M, Rogers GC, Holland G, Scholey JM. Functional coordination of three mitotic motors in Drosophila embryos. Mol Biol Cell. 2000;11:241–253. doi: 10.1091/mbc.11.1.241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Straight AF, Sedat JW, Murray AW. Time-lapse microscopy reveals unique roles for kinesins during anaphase in budding yeast. J Cell Biol. 1998;143:687–694. doi: 10.1083/jcb.143.3.687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi M, Mukai H, Oishi K, Isagawa T, Ono Y. Association of immature hypophosphorylated protein kinase cepsilon with an anchoring protein CG-NAP. J Biol Chem. 2000;275:34592–34596. doi: 10.1074/jbc.M005285200. [DOI] [PubMed] [Google Scholar]
- Takahashi M, Shibata H, Shimakawa M, Miyamoto M, Mukai H, Ono Y. Characterization of a novel giant scaffolding protein, CG-NAP, that anchors multiple signaling enzymes to centrosome and the golgi apparatus. J Biol Chem. 1999;274:17267–17274. doi: 10.1074/jbc.274.24.17267. [DOI] [PubMed] [Google Scholar]
- Takahashi M, Yamagiwa A, Nishimura T, Mukai H, Ono Y. Centrosomal proteins CG-NAP and kendrin provide microtubule nucleation sites by anchoring gamma-tubulin ring complex. Mol Biol Cell. 2002;13:3235–3245. doi: 10.1091/mbc.E02-02-0112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uehara R, Goshima G. Functional central spindle assembly requires de novo microtubule generation in the interchromosomal region during anaphase. J Cell Biol. 2010;191:259–267. doi: 10.1083/jcb.201004150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uehara R, Nozawa R-S, Tomioka A, Petry S, Vale RD, Obuse C, Goshima G. The augmin complex plays a critical role in spindle microtubule generation for mitotic progression and cytokinesis in human cells. Proc Natl Acad Sci USA. 2009;106:6998–7003. doi: 10.1073/pnas.0901587106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uzbekov RE, Votchal MS, Vorobjev IA. Role of the centrosome in mitosis: UV micro-irradiation study. J Photochem Photobiol B. 1995;29:163–170. doi: 10.1016/1011-1344(95)07129-p. [DOI] [PubMed] [Google Scholar]
- Wakefield JG, Huang JY, Raff JW. Centrosomes have a role in regulating the destruction of cyclin B in early Drosophila embryos. Curr Biol. 2000;10:1367–1370. doi: 10.1016/s0960-9822(00)00776-4. [DOI] [PubMed] [Google Scholar]
- Walczak CE, Vernos I, Mitchison TJ, Karsenti E, Heald R. A model for the proposed roles of different microtubule-based motor proteins in establishing spindle bipolarity. Curr Biol. 1998;8:903–913. doi: 10.1016/s0960-9822(07)00370-3. [DOI] [PubMed] [Google Scholar]
- Wiese C, Wilde A, Moore MS, Adam SA, Merdes A, Zheng Y. Role of importin-beta in coupling Ran to downstream targets in microtubule assembly. Science. 2001;291:653–656. doi: 10.1126/science.1057661. [DOI] [PubMed] [Google Scholar]
- Wilde A, Lizarraga SB, Zhang L, Wiese C, Gliksman NR, Walczak CE, Zheng Y. Ran stimulates spindle assembly by altering microtubule dynamics and the balance of motor activities. Nat Cell Biol. 2001;3:221–227. doi: 10.1038/35060000. [DOI] [PubMed] [Google Scholar]
- Wilde A, Zheng Y. Stimulation of microtubule aster formation and spindle assembly by the small GTPase Ran. Science. 1999;284:1359–1362. doi: 10.1126/science.284.5418.1359. [DOI] [PubMed] [Google Scholar]
- Zhang C, Hughes M, Clarke PR. Ran-GTP stabilises microtubule asters and inhibits nuclear assembly in Xenopus egg extracts. J Cell Sci. 1999;112:2453–2461. doi: 10.1242/jcs.112.14.2453. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.