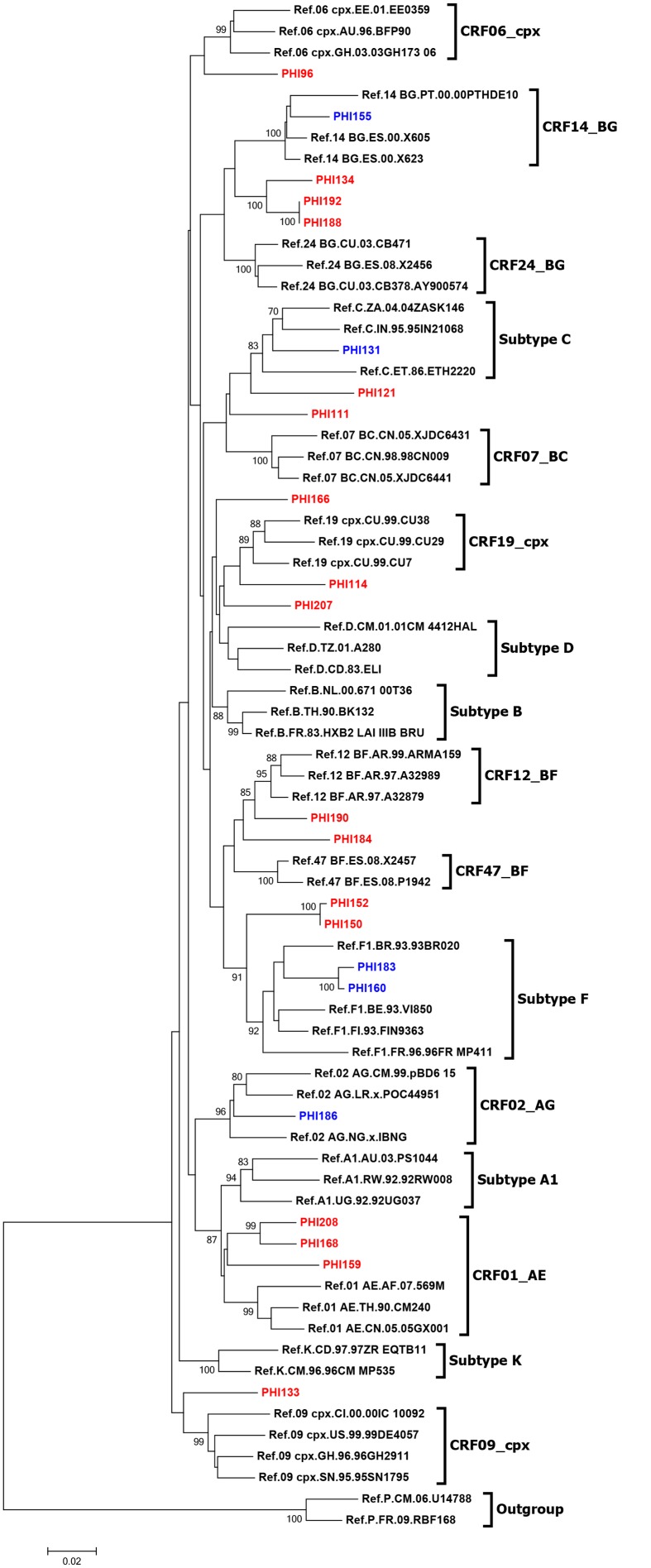
Fig 2. Phylogenetic tree analysis of the non-B HIV-1 pol sequences.
Footnote figure: The phylogenetic inferences were performed by the Neighbor-Joining algorithm under the Kimura-2 parameter nucleotide substitution model with bootstrap. HIV-1 pure subtype and CRFs reference sequences were obtained from Los Alamos National Laboratory (LANL; www.hiv.lanl.gov). Bootstrap values of 70% or greater provide reasonable confidence for genotype assignment (sequences represented in blue). Sequences represented in red were suspected to be recombinants and re-analyzed by bootscanning (data not shown).