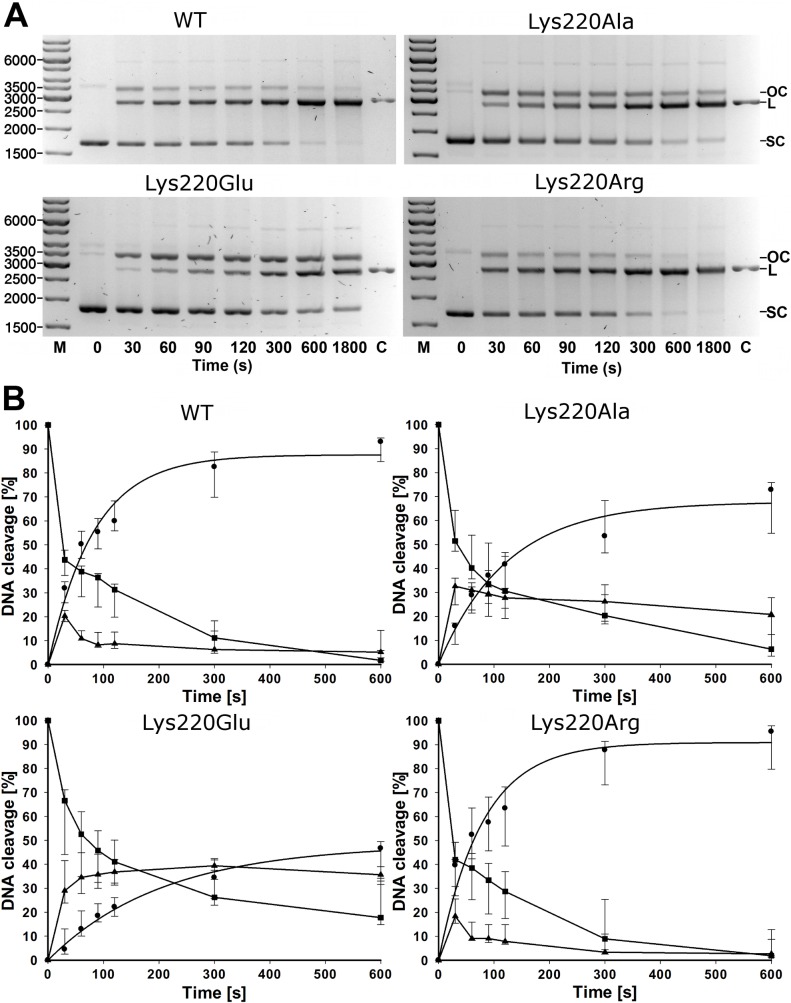
Fig 2. Cleavage of circular DNA.
Circular plasmid DNA bearing one EcoR124I recognition site was reacted with enzymes reconstituted from HsdS1HsdM2 methylase and WT HsdR or mutant HsdRs Lys220Glu, Lys220Ala, or Lys220Arg, and analyzed as in Fig 2. OC, open circular product (▲); L, linear product (●); SC, supercoiled substrate (■); C, control plasmid linearized with HindIII. A: Reactions stopped at the indicated time points were applied to 1.2% agarose gels and visualized by ethidium bromide staining. M is the marker of the indicated numbers of basepairs and C is the linearized plasmind DNA as a control B: Quantification. The three indicated DNA species were quantified individually. Plots for the increase of linear DNA product were derived by fitting an exponential rise to maximum function in SigmaPlot. The points are given for quantification of the gels shown in A, and standard deviations are given from the mean of seven repetitions (WT, Lys220Ala, Lys220Glu) or six repetitions (Lys220Arg) of the experiment conducted with independently purified enzyme preparations.