Fig. 5.
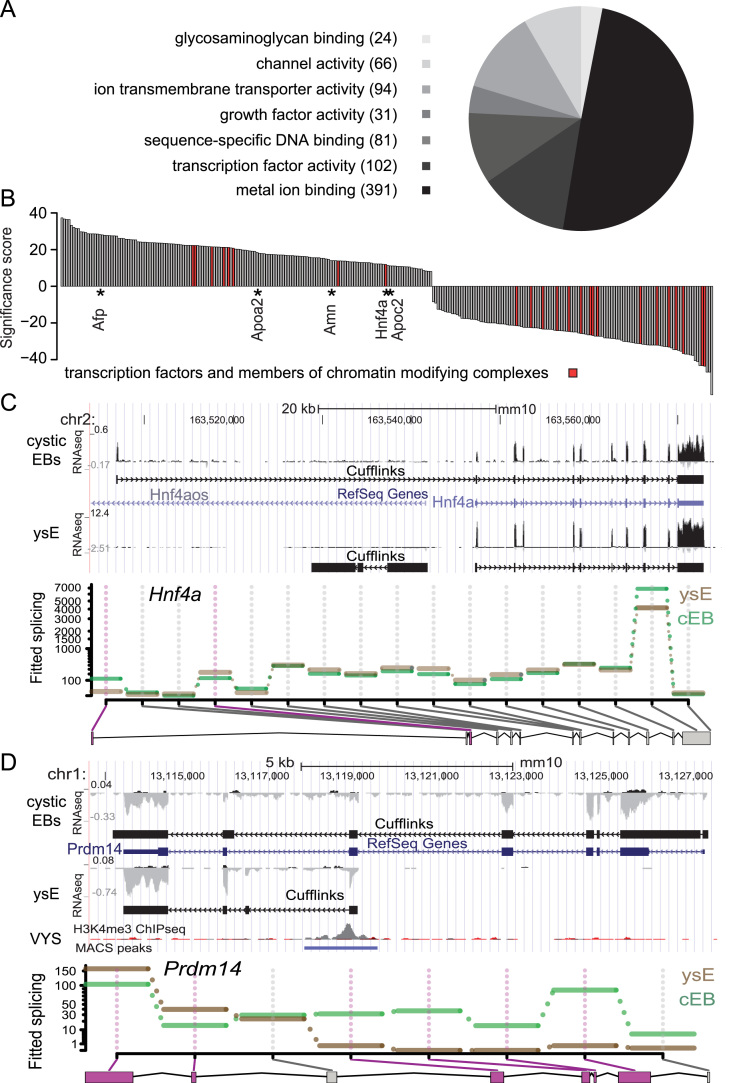
Cystic EBs have a different regulatory network than the yolk sac endoderm. (A) Pie chart representation of gene ontology (GO) analysis of differentially expressed genes between E12.5 ysE and cystic EBs. Significantly differentially expressed genes detected with a FDR of 1% were analyzed using DAVID (Huang et al., 2009). Over represented GO groups/terms (based on an EASE score <10−3) are listed. (B) Genes that show significant differential expression between both developmental stages of ysE (E9.25 and E12.5) and cystic EBs and E12.5 liver are plotted (272 genes, FDR<5%, Fig. S5E). To indicate the direction of differential expression, genes expressed higher in E12.5 ysE are displayed as positive (156 genes) and higher in cystic EBs as negative (118 genes) log2 P values (Table S1). Members of chromatin modifying complexes and transcription factors are highlighted in red. Transcripts that were previously published as markers of ysE are marked with an asterisk. (C) E12.5 ysE expresses the RefSeq annotated shorter isoform of the endoderm specific regulator Hnf4a, while cystic EBs use an alternative promoter to express a longer form that includes a new first exon. Top: UCSC genome browser screenshot showing the RNA-seq tracks and de novo annotation using Cufflinks in cystic EBs and E12.5 ysE (black). Bottom: DEXSeq differential exon usage analysis shows significant differential first exon usage. (D) A novel isoform of the Prdm14 transcription factor distinguishes cystic EBs and E12.5 ysE. Top: A UCSC genome browser screenshot showing RNA-seq for cystic EBs and E12.5 ysE, Cufflinks de novo annotation for both tissues (black), H3K4me3 ChIP-seq for E12.5 VYS (gray, input red) and significant H3K4me3 peaks called using the MACS program. The novel Prdm14 isoform in ysE is supported by a H3K4me3 peak in VYS indicating a novel promoter for this gene. Bottom: DEXSeq differential exon usage analysis shows a significant differential isoform expression between E12.5 ysE and cystic EBs.