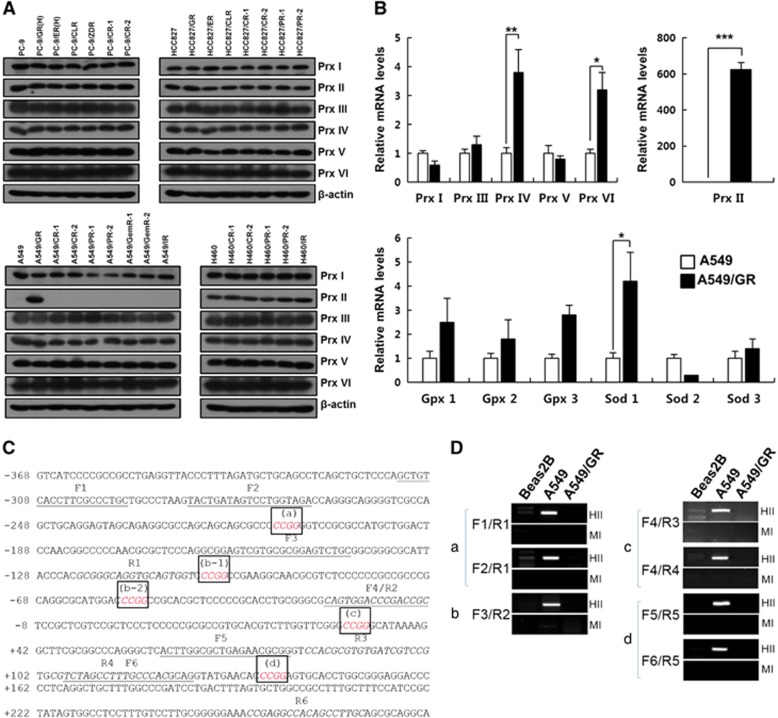
Figure 1.
Expression of antioxidant enzymes and DNA methylation state at peroxiredoxin II (Prx II) promoter in NSCLC cells. (A) Western blot analysis of Prxs in A549 gefitinib- and EGFR-TKI-resistant cells. (B) Quantitative reverse transcriptase-PCR analysis of Prxs and other antioxidant enzymes in A549/GR cells. (C) Sequence information of the Prx II proximal promoter region used for DNA methylation analysis. The HpaII/MspI recognition sequence (5′-CCGG-3′) is italicized and boxed (a, b-1, b-2, c and d). The forward primers (F1–F6) are underlined, and the reverse primers (R1–R6) are italicized. Transcription start site, +1. (D) DNA methylation analysis using restriction enzyme digestion. Genomic DNA was extracted from the indicated cell lines and digested with either HpaII (HII) or MspI (MI). Both enzymes recognize 5′-CCGG-3′, but HpaII is unable to cut DNA when the internal cytosine is methylated. The combinations of primers used in amplifying each of the HpaII/MspI sites (a–d) are indicated. The data are the mean±s.e.m. (n=16) *P<0.05, **P<0.01 and ***P<0.001.