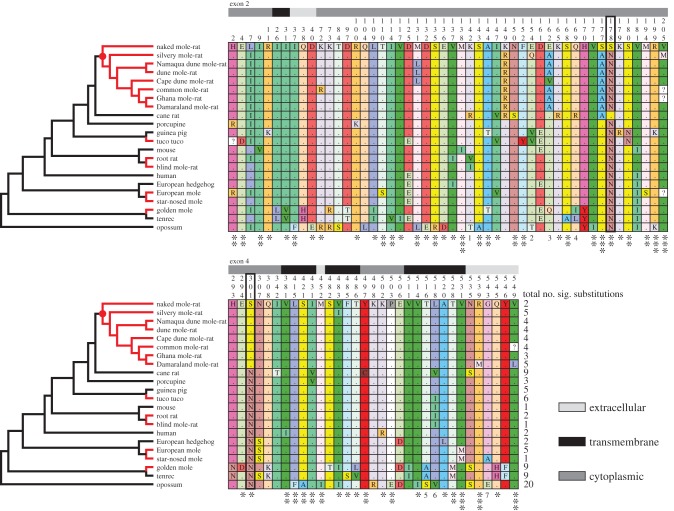
Figure 1.
Phylogenetic relationships and corresponding HAS2 sequences of five clades containing subterranean mammals (red branches), including non-subterranean ingroup comparisons, the human and a marsupial (opossum) outgroup (black branches). The Bathyergidae are the monophyletic clade denoted by the red circle. Adjacent panels show the respective variable amino acids for exons 2 and 4 (site numbers indicated above columns). Shaded bars indicate the relative locations of sites in the molecule (extracellular, transmembrane or cytoplasmic). The key amino acid residues at sites 178 and 301—that facilitate production of HMM-HA in NMRs—are indicated by the bold border. Asterisks below sites denote significance of substitutions estimated by MAPP analysis: *p < 0.05; **p < 0.01; ***p < 0.001; ns, not significant; 1: R = ns, V = **, I = *, T = **; 2: T = **, V = *, Q = ns; 3: V = ***, A = **, Q = ns; 4: R = **, I = ns, L = ns; 5:A = *, S = ns; 6: V = *, I = ns; 7: E = *, A = ns. Numbers at the end of the respective sequence alignments denote the number of substitutions per taxon that are predicted to have a significant impact on protein function.