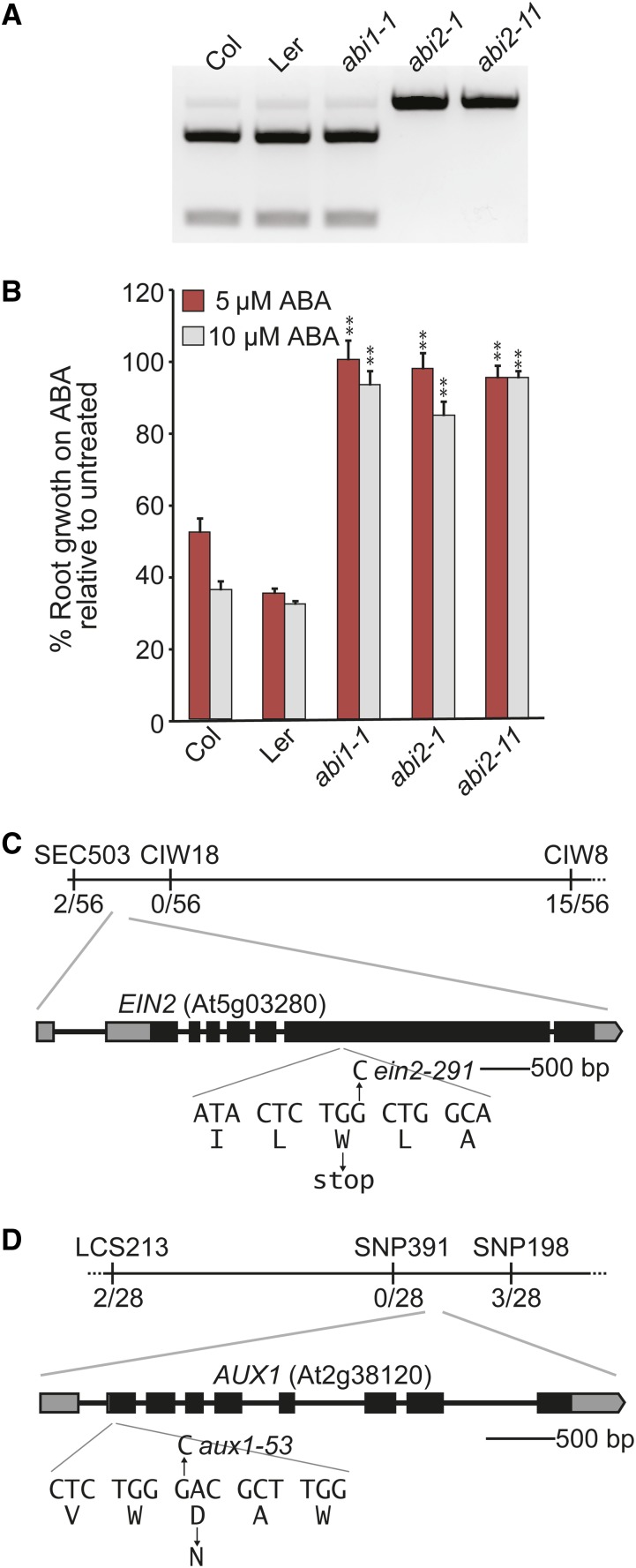
Figure 2.
Identification of AR mutant defects in ABA, ethylene, and auxin response factors. (A) Amplification of ABI2 from wild-type ecotypes Col-0 and Ler-0, abi1-1, abi2-1, and abi2-11, followed by an NcoI digest, demonstrates that abi2-1 and abi2-11 contain the same mutation that removes the NcoI restriction site from the sequence. (B) Mean normalized primary root lengths (±SE; n = 15) of Col-0, Ler-0, abi1-1, abi2-1, and abi2-11 seedlings grown under yellow-filtered light at 22° for 4 d on unsupplemented medium, followed by 4 d on medium supplemented with 5 or 10 µM ABA compared to seedlings grown on medium supplemented with ethanol (mock). Asterisks indicate that the mutant normalized root lengths were significantly longer than wild-type normalized root lengths (**P ≤ 0.001) in two-tailed t tests assuming unequal variance. (C) Recombination mapping with PCR-based markers SEC503, CIW18, and CIW8 localized the ABA resistance mutation in AR291 between SEC503 and CIW8 with 2/56 north and 15/56 south recombinants. Examination of the EIN2 (At5g03280) gene in this region revealed a G-to-A mutation at position 1881 in AR291 DNA that results in Trp455-to-stop. (D) Recombination mapping with PCR-based markers LCS213, SNP391, and SNP198 localized the ABA resistance mutation in AR53 between LCS213 and SNP198 with 2/28 north and 3/28 south recombinants. Examination of the AUX1 (At2g38120) gene in this region revealed a G-to-A mutation at position 148 in AR53 DNA that results in an Asp50-to-Arg substitution.