Figure 4.
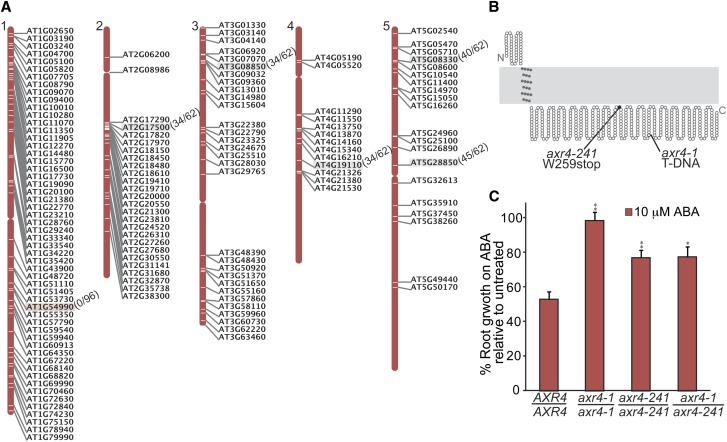
Whole genome sequencing of AR241 revealed a new allele of axr4. (A) Map positions of homozygous EMS-caused mutations identified in AR241. Genomic DNA from pooled M4 seedlings was sequenced and examined for G/C to A/T transitions typically associated with EMS mutagenesis in splice sites and coding sequences (see Table S1 for list of mutations identified). Approximate map positions of the identified mutations are shown to the right of each chromosome. PCR-based markers (Table S2) were designed for a subset of identified mutations and used to localize the AR241 causative mutation near At1g54990 with 0/96 recombination events in a population of AR241 backcrossed lines. Recombination events in examined backcrossed lines are listed to the right of gene names for each PCR-based marker. (B) Examination of AXR4/At1g54990 in AR241 revealed a G-to-A mutation in the first exon causing a Trp259-to-stop mutation. (C) axr4-241 is allelic to axr4-1. Complementation test showing mean normalized primary root lengths (±SE; n ≥ 10) of Col-0 wild-type (AXR4/AXR4), axr4-1/axr4-1, axr4-241/axr4-241, and axr4-1/axr4-241 seedlings grown under yellow-filtered light at 22° for 4 d on unsupplemented medium, followed by 4 d on medium supplemented with 10 µM ABA. Asterisks indicate that the mutant normalized root lengths were significantly longer than wild-type normalized root lengths on ABA (*P ≤ 0.01; **P ≤ 0.001) in two-tailed t tests assuming unequal variance.